Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Evx2
Z-value: 0.43
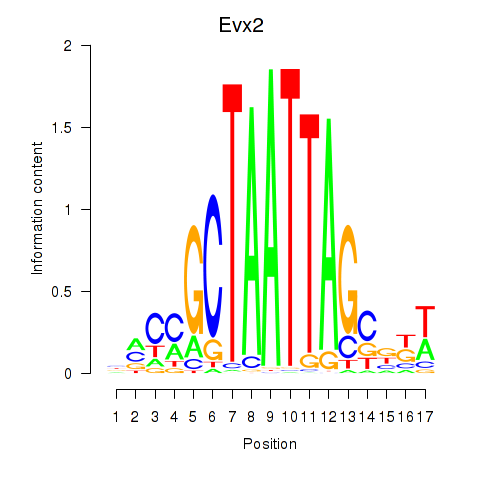
Transcription factors associated with Evx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Evx2
|
ENSRNOG00000001589 | even-skipped homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Evx2 | rn6_v1_chr3_-_61581598_61581598 | 0.47 | 5.1e-19 | Click! |
Activity profile of Evx2 motif
Sorted Z-values of Evx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_28485619 | 18.29 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr11_+_58624198 | 15.57 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr12_-_19167015 | 14.16 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr1_+_59156251 | 13.69 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr8_-_45137893 | 13.13 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr1_-_227457629 | 11.98 |
ENSRNOT00000035910
ENSRNOT00000073770 |
Ms4a5
|
membrane spanning 4-domains A5 |
| chr2_+_66940057 | 11.21 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr12_-_2174131 | 10.99 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr7_-_113228439 | 10.31 |
ENSRNOT00000034672
|
Col22a1
|
collagen type XXII alpha 1 chain |
| chr9_-_30844199 | 9.96 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr1_-_189238776 | 9.79 |
ENSRNOT00000020817
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr3_-_120076788 | 9.59 |
ENSRNOT00000047158
|
Kcnip3
|
potassium voltage-gated channel interacting protein 3 |
| chr10_-_88670430 | 9.33 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr4_+_70977556 | 7.88 |
ENSRNOT00000031984
|
LOC680112
|
hypothetical protein LOC680112 |
| chr2_-_132301073 | 7.57 |
ENSRNOT00000058288
|
RGD1563562
|
similar to GTPase activating protein testicular GAP1 |
| chr13_+_89480058 | 7.48 |
ENSRNOT00000050804
|
Cfap126
|
cilia and flagella associated protein 126 |
| chr1_-_215033460 | 7.45 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr12_-_35979193 | 7.08 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr14_-_115052450 | 7.05 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr11_-_61234944 | 6.68 |
ENSRNOT00000059680
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr7_-_76488216 | 6.47 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr16_+_12955826 | 6.37 |
ENSRNOT00000060751
|
4930596D02Rik
|
RIKEN cDNA 4930596D02 gene |
| chr16_+_51748970 | 6.22 |
ENSRNOT00000059182
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A |
| chr1_+_129613497 | 6.13 |
ENSRNOT00000074410
|
LOC102551255
|
tetraspanin-7-like |
| chr1_+_98414226 | 5.91 |
ENSRNOT00000090785
|
Siglecl1
|
SIGLEC family like 1 |
| chr18_+_4084228 | 5.77 |
ENSRNOT00000071392
|
Cabyr
|
calcium binding tyrosine phosphorylation regulated |
| chr3_+_56056925 | 5.53 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chr5_+_61474000 | 5.31 |
ENSRNOT00000013930
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr18_+_30017918 | 5.23 |
ENSRNOT00000079794
|
Pcdha4
|
protocadherin alpha 4 |
| chr3_+_113319456 | 5.13 |
ENSRNOT00000051354
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr1_-_81550598 | 4.33 |
ENSRNOT00000051671
|
RGD1564380
|
similar to BC049730 protein |
| chr13_+_51229890 | 3.68 |
ENSRNOT00000060669
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr7_-_28711761 | 3.67 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr3_+_80072489 | 3.40 |
ENSRNOT00000057176
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chrX_-_134866210 | 3.35 |
ENSRNOT00000005331
|
Apln
|
apelin |
| chr18_-_16543992 | 3.21 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr3_-_124879216 | 3.04 |
ENSRNOT00000028886
|
Tmem230
|
transmembrane protein 230 |
| chr8_-_83280888 | 3.02 |
ENSRNOT00000052341
|
Gfral
|
GDNF family receptor alpha like |
| chr12_+_41486076 | 2.79 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr7_+_6644643 | 2.78 |
ENSRNOT00000051670
|
Olr962
|
olfactory receptor 962 |
| chr1_+_185863043 | 2.54 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr13_+_90514336 | 2.39 |
ENSRNOT00000088996
ENSRNOT00000085377 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr8_+_126975833 | 2.23 |
ENSRNOT00000088348
|
AABR07071701.1
|
|
| chr19_+_45938915 | 2.20 |
ENSRNOT00000065508
|
Mon1b
|
MON1 homolog B, secretory trafficking associated |
| chr1_+_99888297 | 1.99 |
ENSRNOT00000080896
|
AABR07003241.3
|
|
| chr8_+_57983556 | 1.94 |
ENSRNOT00000009562
|
RGD1311251
|
similar to RIKEN cDNA 4930550C14 |
| chr2_-_219262901 | 1.91 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr5_+_117583502 | 1.78 |
ENSRNOT00000010933
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr10_-_43931094 | 1.73 |
ENSRNOT00000039778
|
Olr1415
|
olfactory receptor 1415 |
| chr2_+_252090669 | 1.69 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr1_-_261179790 | 1.58 |
ENSRNOT00000074420
ENSRNOT00000072073 |
Exosc1
|
exosome component 1 |
| chr7_-_143967484 | 1.53 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr5_-_151473750 | 1.45 |
ENSRNOT00000011357
|
Tmem222
|
transmembrane protein 222 |
| chr1_+_261180007 | 1.43 |
ENSRNOT00000072181
|
Zdhhc16
|
zinc finger, DHHC-type containing 16 |
| chrX_+_110016995 | 1.41 |
ENSRNOT00000093542
|
Nrk
|
Nik related kinase |
| chr17_-_52477575 | 1.40 |
ENSRNOT00000081290
|
Gli3
|
GLI family zinc finger 3 |
| chr1_-_88657845 | 1.40 |
ENSRNOT00000074211
|
LOC100912380
|
calpain small subunit 1-like |
| chr17_-_34945317 | 1.18 |
ENSRNOT00000090457
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr14_+_77922045 | 1.11 |
ENSRNOT00000081198
|
Stk32b
|
serine/threonine kinase 32B |
| chr17_-_87826421 | 1.03 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr15_-_12889102 | 0.91 |
ENSRNOT00000061214
ENSRNOT00000084687 |
RGD1306063
|
similar to HT021 |
| chr20_-_323703 | 0.88 |
ENSRNOT00000089844
|
Olr1668
|
olfactory receptor 1668 |
| chr4_+_71836677 | 0.88 |
ENSRNOT00000024048
|
Tas2r126
|
taste receptor, type 2, member 126 |
| chrX_+_15113878 | 0.67 |
ENSRNOT00000007464
|
Wdr13
|
WD repeat domain 13 |
| chr15_-_93748742 | 0.65 |
ENSRNOT00000093370
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr3_-_133131192 | 0.56 |
ENSRNOT00000055630
|
Tasp1
|
taspase 1 |
| chr13_-_73638073 | 0.54 |
ENSRNOT00000005195
|
Cep350
|
centrosomal protein 350 |
| chr10_-_10881844 | 0.53 |
ENSRNOT00000087118
ENSRNOT00000004416 |
Mgrn1
|
mahogunin ring finger 1 |
| chrX_-_65102344 | 0.52 |
ENSRNOT00000016042
ENSRNOT00000075875 |
Las1l
|
LAS1-like, ribosome biogenesis factor |
| chr20_+_7409401 | 0.45 |
ENSRNOT00000000586
|
Snrpc
|
small nuclear ribonucleoprotein polypeptide C |
| chrX_+_140878216 | 0.45 |
ENSRNOT00000075840
ENSRNOT00000077026 |
Zic3
|
Zic family member 3 |
| chr12_-_48627297 | 0.45 |
ENSRNOT00000000890
|
Iscu
|
iron-sulfur cluster assembly enzyme |
| chr17_+_9797907 | 0.37 |
ENSRNOT00000021638
|
Lman2
|
lectin, mannose-binding 2 |
| chr8_-_19773243 | 0.33 |
ENSRNOT00000040848
|
Olr1155
|
olfactory receptor 1155 |
| chr17_-_27279260 | 0.32 |
ENSRNOT00000018508
|
Snrnp48
|
small nuclear ribonucleoprotein U11/U12 subunit 48 |
| chr15_-_12129220 | 0.30 |
ENSRNOT00000029318
|
Olr1605
|
olfactory receptor 1605 |
| chr16_-_70242979 | 0.30 |
ENSRNOT00000071269
|
LOC103689964
|
vacuolar ATPase assembly integral membrane protein Vma21 |
| chr20_+_12944786 | 0.28 |
ENSRNOT00000048218
|
Pcnt
|
pericentrin |
| chr8_+_107495366 | 0.25 |
ENSRNOT00000056590
|
Cep70
|
centrosomal protein 70 |
| chr13_+_90943255 | 0.21 |
ENSRNOT00000011539
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr1_+_189432604 | 0.17 |
ENSRNOT00000034610
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Evx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 15.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.4 | 11.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 1.2 | 9.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.9 | 18.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.8 | 3.3 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.7 | 2.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.7 | 9.8 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.5 | 2.4 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.4 | 2.5 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.4 | 13.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.4 | 1.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.4 | 3.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.4 | 1.4 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.3 | 9.6 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.3 | 3.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.3 | 6.7 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.3 | 7.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 1.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 1.4 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.2 | 3.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.8 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 1.5 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 16.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 14.2 | GO:0042552 | myelination(GO:0042552) |
| 0.1 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 2.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.5 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 3.0 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 3.7 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 0.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.7 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 6.5 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 12.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.6 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 1.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 7.5 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 2.2 | GO:0097480 | synaptic vesicle transport(GO:0048489) establishment of synaptic vesicle localization(GO:0097480) |
| 0.0 | 0.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 15.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.7 | 14.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 5.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 3.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 9.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 9.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 5.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 7.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 2.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 3.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 11.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 5.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 3.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 11.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.5 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 15.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 1.5 | 18.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 1.2 | 9.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.8 | 1.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 5.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 3.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.5 | 7.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 2.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.3 | 12.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.3 | 3.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 6.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 9.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.5 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 3.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.5 | GO:0030619 | U1 snRNA binding(GO:0030619) U1 snRNP binding(GO:1990446) |
| 0.0 | 11.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 17.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 13.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 3.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.4 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 15.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 15.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 7.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 11.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 20.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 1.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 3.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 11.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |