Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Etv6
Z-value: 0.82
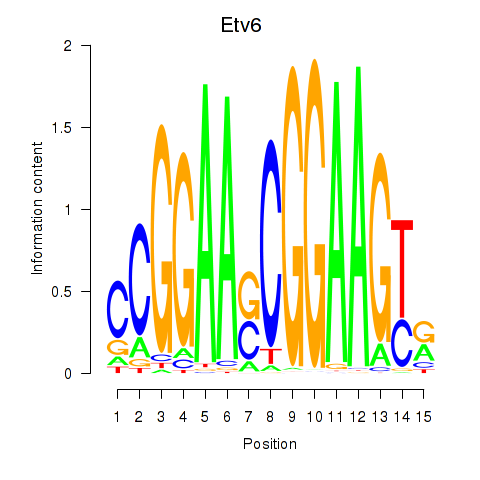
Transcription factors associated with Etv6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv6
|
ENSRNOG00000005984 | ets variant 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Etv6 | rn6_v1_chr4_+_167754525_167754525 | -0.09 | 1.1e-01 | Click! |
Activity profile of Etv6 motif
Sorted Z-values of Etv6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_105355716 | 11.15 |
ENSRNOT00000015043
|
Timm8a1
|
translocase of inner mitochondrial membrane 8 homolog A1 (yeast) |
| chr8_-_87213627 | 10.52 |
ENSRNOT00000066084
|
Cox7a2
|
cytochrome c oxidase subunit 7A2 |
| chr3_-_79728879 | 8.73 |
ENSRNOT00000012425
|
Ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3 |
| chr19_+_52032886 | 6.78 |
ENSRNOT00000019923
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr10_-_20658100 | 6.54 |
ENSRNOT00000010252
|
Rars
|
arginyl-tRNA synthetase |
| chr1_-_199395363 | 6.24 |
ENSRNOT00000090368
ENSRNOT00000026587 |
Prss36
|
protease, serine, 36 |
| chr4_-_153373649 | 6.11 |
ENSRNOT00000016495
|
Atp6v1e1
|
ATPase H+ transporting V1 subunit E1 |
| chr9_-_17254949 | 6.07 |
ENSRNOT00000026499
|
Mrps18a
|
mitochondrial ribosomal protein S18A |
| chr1_-_216953999 | 5.94 |
ENSRNOT00000028126
|
Mrgprg
|
MAS related GPR family member G |
| chr10_+_55924938 | 5.87 |
ENSRNOT00000087003
ENSRNOT00000057079 |
Trappc1
|
trafficking protein particle complex 1 |
| chr10_+_75529009 | 5.82 |
ENSRNOT00000013771
|
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr1_-_264756546 | 5.69 |
ENSRNOT00000020020
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr9_-_23352668 | 5.51 |
ENSRNOT00000075279
|
Mut
|
methylmalonyl CoA mutase |
| chr7_+_144647587 | 5.43 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr10_-_109630005 | 5.30 |
ENSRNOT00000075206
|
Oxld1
|
oxidoreductase-like domain containing 1 |
| chr1_-_220746224 | 5.19 |
ENSRNOT00000027734
|
Banf1
|
barrier to autointegration factor 1 |
| chr13_-_97838228 | 5.14 |
ENSRNOT00000003618
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr7_+_97760134 | 4.96 |
ENSRNOT00000086792
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr12_+_52467450 | 4.86 |
ENSRNOT00000056675
|
Pgam5
|
PGAM family member 5, mitochondrial serine/threonine protein phosphatase |
| chr17_+_72209373 | 4.85 |
ENSRNOT00000064802
|
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr1_+_225068009 | 4.78 |
ENSRNOT00000026651
|
Ubxn1
|
UBX domain protein 1 |
| chr5_+_137670295 | 4.65 |
ENSRNOT00000076672
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr10_+_75032365 | 4.49 |
ENSRNOT00000010448
|
Supt4h1
|
SPT4 homolog, DSIF elongation factor subunit |
| chr2_-_189096785 | 4.47 |
ENSRNOT00000028200
|
Chrnb2
|
cholinergic receptor nicotinic beta 2 subunit |
| chr9_-_16581078 | 4.44 |
ENSRNOT00000022582
|
Pex6
|
peroxisomal biogenesis factor 6 |
| chr16_-_7082193 | 4.44 |
ENSRNOT00000024447
|
Spcs1
|
signal peptidase complex subunit 1 |
| chr8_+_21663325 | 4.39 |
ENSRNOT00000027749
|
Ubl5
|
ubiquitin-like 5 |
| chr2_-_197982385 | 4.34 |
ENSRNOT00000034074
|
Mrps21
|
mitochondrial ribosomal protein S21 |
| chr5_+_137458691 | 4.29 |
ENSRNOT00000075464
ENSRNOT00000076895 |
LOC108348080
|
probable rRNA-processing protein EBP2 |
| chr9_+_65478496 | 4.26 |
ENSRNOT00000016060
|
Ndufb3
|
NADH:ubiquinone oxidoreductase subunit B3 |
| chr3_+_125470551 | 4.21 |
ENSRNOT00000028898
|
Mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr20_+_22728208 | 4.17 |
ENSRNOT00000000794
|
LOC100363472
|
nuclear receptor-binding factor 2-like |
| chr9_+_81672758 | 4.12 |
ENSRNOT00000020646
|
Ctdsp1
|
CTD small phosphatase 1 |
| chr8_-_22173797 | 4.07 |
ENSRNOT00000042314
ENSRNOT00000079949 |
Cdc37
|
cell division cycle 37 |
| chr1_+_218532045 | 4.04 |
ENSRNOT00000018848
|
Mrpl21
|
mitochondrial ribosomal protein L21 |
| chr20_+_13760810 | 4.04 |
ENSRNOT00000081140
ENSRNOT00000080203 |
Gstt2
|
glutathione S-transferase, theta 2 |
| chr2_-_196046311 | 4.03 |
ENSRNOT00000028484
|
Psmb4
|
proteasome subunit beta 4 |
| chr7_-_121783435 | 3.98 |
ENSRNOT00000034912
|
Enthd1
|
ENTH domain containing 1 |
| chr5_+_137670123 | 3.87 |
ENSRNOT00000072245
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr1_+_266358728 | 3.85 |
ENSRNOT00000027118
|
Wbp1l
|
WW domain binding protein 1-like |
| chrX_+_157095937 | 3.82 |
ENSRNOT00000091792
|
Bcap31
|
B-cell receptor-associated protein 31 |
| chr8_-_21661259 | 3.81 |
ENSRNOT00000068308
|
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chrX_+_27015884 | 3.81 |
ENSRNOT00000065814
|
Msl3
|
male-specific lethal 3 homolog (Drosophila) |
| chr1_-_216942782 | 3.80 |
ENSRNOT00000049057
|
RGD1562011
|
similar to MAS-related G-protein coupled receptor, member G |
| chr1_-_36112993 | 3.75 |
ENSRNOT00000023161
|
Med10
|
mediator complex subunit 10 |
| chr6_+_108936664 | 3.62 |
ENSRNOT00000007298
|
Dlst
|
dihydrolipoamide S-succinyltransferase |
| chr9_+_82718709 | 3.56 |
ENSRNOT00000027256
ENSRNOT00000080524 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr3_-_12007570 | 3.50 |
ENSRNOT00000060186
|
Lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr19_-_41433346 | 3.49 |
ENSRNOT00000022952
|
Cmtr2
|
cap methyltransferase 2 |
| chr17_-_29438668 | 3.42 |
ENSRNOT00000021660
|
Fars2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr19_+_45938915 | 3.40 |
ENSRNOT00000065508
|
Mon1b
|
MON1 homolog B, secretory trafficking associated |
| chr8_+_70522092 | 3.31 |
ENSRNOT00000025873
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr3_+_147713821 | 3.31 |
ENSRNOT00000007558
|
Csnk2a1
|
casein kinase 2 alpha 1 |
| chr15_+_24078280 | 3.27 |
ENSRNOT00000015511
ENSRNOT00000063807 |
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr12_+_8976585 | 3.27 |
ENSRNOT00000071796
|
LOC100911238
|
proteasome maturation protein-like |
| chr10_-_70516421 | 3.26 |
ENSRNOT00000013233
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr12_-_6784111 | 3.22 |
ENSRNOT00000078541
|
AABR07035203.1
|
|
| chr12_+_16899025 | 3.21 |
ENSRNOT00000001716
|
Psmg3
|
proteasome assembly chaperone 3 |
| chr2_+_54897424 | 3.12 |
ENSRNOT00000017665
|
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr10_+_3321476 | 3.11 |
ENSRNOT00000090166
|
Mpv17l
|
MPV17 mitochondrial inner membrane protein like |
| chr5_+_142332607 | 3.10 |
ENSRNOT00000072841
|
LOC100909856
|
metal regulatory transcription factor 1-like |
| chr3_-_125470413 | 3.10 |
ENSRNOT00000028893
|
Trmt6
|
tRNA methyltransferase 6 |
| chr15_+_28287024 | 3.07 |
ENSRNOT00000064907
|
Mettl17
|
methyltransferase like 17 |
| chr3_+_151032952 | 3.06 |
ENSRNOT00000064013
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr19_+_34041219 | 2.97 |
ENSRNOT00000017226
|
Tmem184c
|
transmembrane protein 184C |
| chr10_+_82375572 | 2.96 |
ENSRNOT00000004947
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr5_-_14588422 | 2.91 |
ENSRNOT00000011312
|
Lypla1
|
lysophospholipase I |
| chr3_-_119405300 | 2.90 |
ENSRNOT00000015568
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chr6_+_75996643 | 2.87 |
ENSRNOT00000052076
|
Srp54a
|
signal recognition particle 54A |
| chr9_+_111220858 | 2.87 |
ENSRNOT00000076669
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr5_-_70463546 | 2.86 |
ENSRNOT00000043184
|
AC118841.1
|
|
| chr1_+_226947105 | 2.84 |
ENSRNOT00000028373
|
Prpf19
|
pre-mRNA processing factor 19 |
| chr5_+_48047913 | 2.79 |
ENSRNOT00000065926
|
Lyrm2
|
LYR motif containing 2 |
| chr12_-_8759599 | 2.78 |
ENSRNOT00000073155
|
Pomp
|
proteasome maturation protein |
| chr12_-_37468935 | 2.69 |
ENSRNOT00000001387
|
Ddx55
|
DEAD-box helicase 55 |
| chr10_+_84976377 | 2.66 |
ENSRNOT00000013114
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr5_-_138222534 | 2.62 |
ENSRNOT00000009691
|
Zfp691
|
zinc finger protein 691 |
| chr15_-_55424024 | 2.59 |
ENSRNOT00000077733
|
Nudt15
|
nudix hydrolase 15 |
| chr7_-_139734568 | 2.58 |
ENSRNOT00000079377
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr6_-_126636484 | 2.56 |
ENSRNOT00000088000
ENSRNOT00000063786 |
Gon7
|
GON7, KEOPS complex subunit homolog |
| chr1_+_264756499 | 2.51 |
ENSRNOT00000031018
|
Peo1
|
progressive external ophthalmoplegia 1 |
| chr11_-_27080701 | 2.50 |
ENSRNOT00000002180
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr3_+_7422820 | 2.48 |
ENSRNOT00000064323
|
Ddx31
|
DEAD-box helicase 31 |
| chr5_+_142797366 | 2.45 |
ENSRNOT00000031928
|
Mtf1
|
metal-regulatory transcription factor 1 |
| chr10_+_109630099 | 2.32 |
ENSRNOT00000072099
|
Ccdc137
|
coiled-coil domain containing 137 |
| chr3_-_140141679 | 2.32 |
ENSRNOT00000014632
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr6_+_126636662 | 2.31 |
ENSRNOT00000010739
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr3_-_7422738 | 2.31 |
ENSRNOT00000088339
|
Gtf3c4
|
general transcription factor IIIC subunit 4 |
| chr17_-_90071408 | 2.30 |
ENSRNOT00000071978
|
Pdss1
|
decaprenyl diphosphate synthase subunit 1 |
| chr7_+_117394372 | 2.30 |
ENSRNOT00000048706
|
Gpaa1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chrX_-_1704033 | 2.25 |
ENSRNOT00000051956
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr9_-_111220651 | 2.24 |
ENSRNOT00000016028
|
Gin1
|
gypsy retrotransposon integrase 1 |
| chr6_-_102047758 | 2.22 |
ENSRNOT00000012101
|
Atp6v1d
|
ATPase H+ transporting V1 subunit D |
| chr8_+_68526093 | 2.20 |
ENSRNOT00000011385
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr5_-_138361702 | 2.19 |
ENSRNOT00000067943
ENSRNOT00000064712 |
Ppih
|
peptidylprolyl isomerase H |
| chr12_+_51937175 | 2.18 |
ENSRNOT00000056780
|
Pus1
|
pseudouridylate synthase 1 |
| chr5_-_172488822 | 2.16 |
ENSRNOT00000019620
|
Rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr3_-_119405453 | 2.13 |
ENSRNOT00000090355
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chr19_-_39646693 | 2.11 |
ENSRNOT00000019104
|
Psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr15_+_34270648 | 2.09 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr17_+_29438980 | 2.09 |
ENSRNOT00000072056
|
Lyrm4
|
LYR motif containing 4 |
| chr10_+_107455845 | 2.08 |
ENSRNOT00000004373
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr16_-_81945127 | 2.07 |
ENSRNOT00000023352
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr10_-_109639054 | 2.05 |
ENSRNOT00000071433
|
Arl16
|
ADP-ribosylation factor like GTPase 16 |
| chr1_+_198911911 | 2.03 |
ENSRNOT00000055002
|
Prr14
|
proline rich 14 |
| chr1_-_215290838 | 2.00 |
ENSRNOT00000028283
ENSRNOT00000047669 |
Cttn
|
cortactin |
| chr7_-_12399910 | 1.99 |
ENSRNOT00000021677
|
RGD1562114
|
RGD1562114 |
| chr7_+_74994605 | 1.97 |
ENSRNOT00000013801
|
Spag1
|
sperm associated antigen 1 |
| chr2_+_188757489 | 1.93 |
ENSRNOT00000028052
|
Pygo2
|
pygopus family PHD finger 2 |
| chr1_-_173682226 | 1.92 |
ENSRNOT00000020137
|
Ric3
|
RIC3 acetylcholine receptor chaperone |
| chr12_-_37398329 | 1.90 |
ENSRNOT00000083401
|
Tctn2
|
tectonic family member 2 |
| chr20_+_3309899 | 1.89 |
ENSRNOT00000001049
|
Abcf1
|
ATP binding cassette subfamily F member 1 |
| chr15_-_86142672 | 1.82 |
ENSRNOT00000057830
|
Commd6
|
COMM domain containing 6 |
| chr4_+_144985880 | 1.79 |
ENSRNOT00000009464
|
Thumpd3
|
THUMP domain containing 3 |
| chr10_+_43768708 | 1.78 |
ENSRNOT00000086218
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr1_-_103128743 | 1.77 |
ENSRNOT00000075006
|
Spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr1_+_29432152 | 1.75 |
ENSRNOT00000019436
ENSRNOT00000083276 |
Trmt11
|
tRNA methyltransferase 11 homolog |
| chr6_+_126636491 | 1.74 |
ENSRNOT00000089601
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr6_-_105261549 | 1.70 |
ENSRNOT00000009122
|
Synj2bp
|
synaptojanin 2 binding protein |
| chr2_-_183582553 | 1.68 |
ENSRNOT00000014236
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr19_+_53055745 | 1.67 |
ENSRNOT00000074430
|
Foxl1
|
forkhead box L1 |
| chr1_+_222250995 | 1.66 |
ENSRNOT00000031458
|
Trpt1
|
tRNA phosphotransferase 1 |
| chr1_-_102970950 | 1.63 |
ENSRNOT00000018194
|
Tsg101
|
tumor susceptibility 101 |
| chr10_-_34990943 | 1.62 |
ENSRNOT00000075555
|
Rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr1_+_143583675 | 1.62 |
ENSRNOT00000026385
|
Fam103a1
|
family with sequence similarity 103, member A1 |
| chr11_-_24294179 | 1.61 |
ENSRNOT00000002116
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chrX_+_115561332 | 1.60 |
ENSRNOT00000076680
ENSRNOT00000075912 ENSRNOT00000007968 |
Alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr1_-_196884302 | 1.59 |
ENSRNOT00000089464
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr3_-_21697945 | 1.57 |
ENSRNOT00000012433
|
Zbtb26
|
zinc finger and BTB domain containing 26 |
| chr13_+_44957014 | 1.55 |
ENSRNOT00000004878
|
Ubxn4
|
UBX domain protein 4 |
| chr7_-_2825498 | 1.54 |
ENSRNOT00000086656
ENSRNOT00000031362 |
Nabp2
|
nucleic acid binding protein 2 |
| chr1_-_101426852 | 1.54 |
ENSRNOT00000028217
|
Ruvbl2
|
RuvB-like AAA ATPase 2 |
| chr1_-_226255886 | 1.53 |
ENSRNOT00000027842
|
Fen1
|
flap structure-specific endonuclease 1 |
| chr10_-_37209881 | 1.51 |
ENSRNOT00000090475
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr16_+_7082450 | 1.49 |
ENSRNOT00000013291
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr7_-_33584564 | 1.47 |
ENSRNOT00000005463
|
Nedd1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr10_-_67285617 | 1.47 |
ENSRNOT00000019044
|
Utp6
|
UTP6 small subunit processome component |
| chr2_+_263864331 | 1.45 |
ENSRNOT00000055259
|
Zranb2
|
zinc finger RANBP2-type containing 2 |
| chr1_+_80056755 | 1.44 |
ENSRNOT00000021221
|
Snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr3_+_21698032 | 1.44 |
ENSRNOT00000013378
|
Rabgap1
|
RAB GTPase activating protein 1 |
| chr10_+_93354003 | 1.43 |
ENSRNOT00000008140
|
Mettl2b
|
methyltransferase like 2B |
| chr18_-_40732126 | 1.42 |
ENSRNOT00000000170
|
Atg12
|
autophagy related 12 |
| chr5_+_47186558 | 1.42 |
ENSRNOT00000007654
|
LOC100910771
|
mitogen-activated protein kinase kinase kinase 7-like |
| chr4_-_59014416 | 1.39 |
ENSRNOT00000049811
|
RGD1561636
|
similar to 60S ribosomal protein L38 |
| chr1_+_147021436 | 1.38 |
ENSRNOT00000042490
|
F8a1
|
coagulation factor VIII-associated 1 |
| chr15_-_34352673 | 1.37 |
ENSRNOT00000064916
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr20_-_5112872 | 1.35 |
ENSRNOT00000076983
ENSRNOT00000001123 ENSRNOT00000076528 |
Csnk2b
|
casein kinase 2 beta |
| chr12_+_23989596 | 1.35 |
ENSRNOT00000001960
|
Tmem120a
|
transmembrane protein 120A |
| chr19_-_39257451 | 1.33 |
ENSRNOT00000066312
|
Cog8
|
component of oligomeric golgi complex 8 |
| chr13_+_85465792 | 1.32 |
ENSRNOT00000082991
ENSRNOT00000005277 |
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr10_-_35716260 | 1.31 |
ENSRNOT00000004308
ENSRNOT00000059255 |
Sqstm1
|
sequestosome 1 |
| chr2_-_62887048 | 1.29 |
ENSRNOT00000039234
|
RGD1306502
|
similar to hypothetical protein FLJ11193 |
| chr9_-_42805673 | 1.23 |
ENSRNOT00000020558
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr10_-_108425206 | 1.22 |
ENSRNOT00000073140
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr6_+_129609375 | 1.20 |
ENSRNOT00000083732
|
Papola
|
poly (A) polymerase alpha |
| chr3_+_110918243 | 1.19 |
ENSRNOT00000056432
|
Rad51
|
RAD51 recombinase |
| chr7_+_12256387 | 1.19 |
ENSRNOT00000021939
|
RGD1359127
|
similar to RIKEN cDNA 2310011J03 |
| chr13_+_56513286 | 1.14 |
ENSRNOT00000015596
|
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr3_+_122947913 | 1.12 |
ENSRNOT00000028821
|
Vps16
|
VPS16 CORVET/HOPS core subunit |
| chr14_-_104375649 | 1.12 |
ENSRNOT00000006607
|
Actr2
|
ARP2 actin related protein 2 homolog |
| chr1_+_60717386 | 1.10 |
ENSRNOT00000015019
|
Ppp2r1a
|
protein phosphatase 2 scaffold subunit A alpha |
| chr4_-_145699577 | 1.10 |
ENSRNOT00000014377
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr9_+_114022137 | 1.08 |
ENSRNOT00000007657
|
Map3k7
|
mitogen activated protein kinase kinase kinase 7 |
| chr6_+_10594122 | 1.07 |
ENSRNOT00000020534
|
Cript
|
CXXC repeat containing interactor of PDZ3 domain |
| chr1_-_217638118 | 1.07 |
ENSRNOT00000054859
ENSRNOT00000092916 |
Cttn
|
cortactin |
| chr3_+_11554457 | 1.06 |
ENSRNOT00000073087
|
Fam102a
|
family with sequence similarity 102, member A |
| chr8_+_49354115 | 1.05 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr10_-_94445503 | 1.04 |
ENSRNOT00000013295
|
Ftsj3
|
FtsJ homolog 3 |
| chr1_+_31700953 | 1.04 |
ENSRNOT00000020251
|
Exoc3
|
exocyst complex component 3 |
| chr13_-_37287458 | 1.03 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr12_+_11240761 | 1.02 |
ENSRNOT00000001310
|
Pdap1
|
PDGFA associated protein 1 |
| chrX_-_70461553 | 1.01 |
ENSRNOT00000076110
ENSRNOT00000003872 ENSRNOT00000076812 |
Pdzd11
|
PDZ domain containing 11 |
| chr10_+_28243 | 1.00 |
ENSRNOT00000003193
ENSRNOT00000087156 |
LOC103689943
|
meiosis arrest female protein 1-like |
| chr4_-_157486844 | 0.99 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr4_-_7113919 | 0.98 |
ENSRNOT00000014082
|
Chpf2
|
chondroitin polymerizing factor 2 |
| chr2_+_189400696 | 0.91 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr3_-_94419048 | 0.91 |
ENSRNOT00000015775
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr10_+_94445877 | 0.90 |
ENSRNOT00000013997
|
Psmc5
|
proteasome 26S subunit, ATPase 5 |
| chr5_+_126226291 | 0.89 |
ENSRNOT00000009582
|
Ttc22
|
tetratricopeptide repeat domain 22 |
| chr1_-_170586603 | 0.86 |
ENSRNOT00000025955
|
Taf10
|
TATA-box binding protein associated factor 10 |
| chr5_-_134348995 | 0.84 |
ENSRNOT00000015961
|
Faah
|
fatty acid amide hydrolase |
| chr6_+_108796182 | 0.84 |
ENSRNOT00000006297
|
Fcf1
|
FCF1 rRNA-processing protein |
| chr1_-_218532118 | 0.83 |
ENSRNOT00000086385
ENSRNOT00000018487 |
Ighmbp2
|
immunoglobulin mu binding protein 2 |
| chr8_-_21968415 | 0.82 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr10_+_48773828 | 0.82 |
ENSRNOT00000004113
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr1_+_65541322 | 0.82 |
ENSRNOT00000030931
|
Chmp2a
|
charged multivesicular body protein 2A |
| chr1_+_197839430 | 0.81 |
ENSRNOT00000025043
|
Rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr10_-_109802739 | 0.80 |
ENSRNOT00000054951
|
Sirt7
|
sirtuin 7 |
| chr19_+_30668602 | 0.79 |
ENSRNOT00000035084
|
Usp38
|
ubiquitin specific peptidase 38 |
| chr1_-_80515694 | 0.79 |
ENSRNOT00000075144
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr19_+_39257586 | 0.78 |
ENSRNOT00000027653
|
Nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr2_-_58534211 | 0.75 |
ENSRNOT00000089178
|
Skp2
|
S-phase kinase associated protein 2 |
| chr2_+_165805144 | 0.72 |
ENSRNOT00000073716
|
Rabif
|
RAB interacting factor |
| chr7_-_2972521 | 0.72 |
ENSRNOT00000061995
|
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr7_-_97759852 | 0.72 |
ENSRNOT00000007484
|
Derl1
|
derlin 1 |
| chr8_-_62542622 | 0.71 |
ENSRNOT00000080071
|
Clk3
|
CDC-like kinase 3 |
| chr10_-_37084637 | 0.70 |
ENSRNOT00000075525
|
LOC103689927
|
protein RMD5 homolog B |
| chr16_+_18975746 | 0.69 |
ENSRNOT00000066121
|
Smim7
|
small integral membrane protein 7 |
| chrX_-_112328642 | 0.68 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr9_-_81880105 | 0.66 |
ENSRNOT00000022677
|
Rnf25
|
ring finger protein 25 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Etv6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0036115 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) fatty-acyl-CoA catabolic process(GO:0036115) |
| 1.8 | 5.4 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 1.6 | 6.5 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 1.5 | 4.5 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 1.3 | 4.0 | GO:0009751 | response to salicylic acid(GO:0009751) |
| 1.3 | 5.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 1.3 | 3.8 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.2 | 7.4 | GO:0015074 | DNA integration(GO:0015074) |
| 1.2 | 3.6 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 1.2 | 4.8 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 1.2 | 3.5 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 1.1 | 3.4 | GO:0019086 | late viral transcription(GO:0019086) |
| 1.1 | 4.4 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 1.0 | 3.1 | GO:0006083 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 0.8 | 3.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.7 | 2.9 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.7 | 2.8 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.6 | 3.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.6 | 1.8 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.6 | 7.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.6 | 1.7 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.5 | 2.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.5 | 2.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.5 | 2.6 | GO:0006203 | dGTP catabolic process(GO:0006203) DNA protection(GO:0042262) |
| 0.5 | 1.5 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.5 | 2.3 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.5 | 5.0 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.4 | 4.5 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.4 | 1.2 | GO:0072714 | response to selenite ion(GO:0072714) regulation of selenocysteine incorporation(GO:1904569) |
| 0.4 | 1.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.4 | 6.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 2.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.4 | 3.1 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.4 | 1.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 4.1 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.3 | 5.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.3 | 1.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.3 | 1.2 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.3 | 6.1 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 1.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 3.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 1.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 2.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.2 | 3.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 6.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 1.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 1.1 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.2 | 1.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 0.8 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 0.8 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.2 | 0.6 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 4.0 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.2 | 14.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.2 | 8.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 9.3 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.2 | 1.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 1.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 | 1.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 0.6 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 4.9 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.2 | 1.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.2 | 2.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 1.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.9 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 2.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.8 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 1.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.4 | GO:0033123 | positive regulation of purine nucleotide catabolic process(GO:0033123) positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.1 | 1.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.7 | GO:1903671 | negative regulation of activin receptor signaling pathway(GO:0032926) negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.1 | 5.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.3 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 8.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 0.9 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.8 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 1.0 | GO:0071501 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 1.5 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.1 | 1.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 1.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 1.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 4.3 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 1.8 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 2.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.8 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.9 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 2.2 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 0.3 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 1.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 1.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 3.3 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 2.2 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 4.1 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 5.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.9 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 0.2 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 0.8 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 1.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 1.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 5.5 | GO:0007292 | female gamete generation(GO:0007292) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 2.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 2.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 1.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 3.7 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 1.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 3.8 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 1.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.8 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 1.5 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 2.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 2.0 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 1.4 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.3 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 1.0 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 2.7 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.8 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 1.1 | 4.5 | GO:0032044 | DSIF complex(GO:0032044) |
| 1.1 | 3.3 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 1.0 | 3.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 1.0 | 3.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.9 | 2.8 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.8 | 3.8 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.7 | 2.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.7 | 3.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.7 | 5.0 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.6 | 3.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.6 | 4.8 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.6 | 2.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.5 | 4.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.5 | 3.1 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.5 | 6.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.5 | 5.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.5 | 13.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.4 | 6.1 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.4 | 2.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 1.6 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.4 | 1.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 2.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.4 | 4.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 10.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.4 | 2.9 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.4 | 1.4 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.3 | 1.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 6.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 2.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.3 | 15.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.3 | 1.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.3 | 13.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 8.5 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.2 | 4.1 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.2 | 1.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 0.7 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.2 | 4.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 1.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 3.7 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.2 | 3.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 11.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 7.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 1.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.9 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 1.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.6 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 1.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.4 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.1 | 1.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 4.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 5.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 4.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 4.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 3.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 6.5 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.6 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 4.9 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 3.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.2 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 3.5 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 2.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.6 | 6.5 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) arginine binding(GO:0034618) |
| 1.2 | 3.6 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 1.2 | 3.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 1.0 | 4.8 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.9 | 2.6 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.8 | 5.0 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.8 | 3.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.7 | 2.9 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.7 | 2.9 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.7 | 5.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.7 | 4.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.7 | 6.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.6 | 1.7 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.5 | 4.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.5 | 3.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.5 | 4.9 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 1.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.4 | 2.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.4 | 2.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) GPI anchor binding(GO:0034235) |
| 0.4 | 1.4 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.3 | 8.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 1.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 0.9 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.3 | 8.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 0.8 | GO:0004040 | amidase activity(GO:0004040) |
| 0.3 | 3.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 4.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 4.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 2.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 4.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 1.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 5.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 4.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 3.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 4.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 1.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.2 | 0.8 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 1.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 0.6 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.2 | 4.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 1.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 1.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 4.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 7.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 1.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 0.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 1.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 2.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 5.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 1.0 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 2.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 21.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 5.2 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 2.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 4.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 3.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 4.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 3.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 2.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 9.8 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 3.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 5.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 6.7 | GO:0042623 | ATPase activity, coupled(GO:0042623) |
| 0.0 | 1.6 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.8 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 1.0 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.8 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 1.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 8.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.5 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.3 | GO:0005262 | calcium channel activity(GO:0005262) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 4.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 0.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.4 | 6.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.4 | 5.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 4.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 3.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 4.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 4.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 8.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 1.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 6.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 3.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.2 | 11.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 1.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 7.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.5 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 2.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.9 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 0.6 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 5.1 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.1 | 1.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 2.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.8 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 3.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 6.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |