Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Etv4
Z-value: 0.65
Transcription factors associated with Etv4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv4
|
ENSRNOG00000020792 | ets variant 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Etv4 | rn6_v1_chr10_-_89700283_89700283 | 0.47 | 4.3e-19 | Click! |
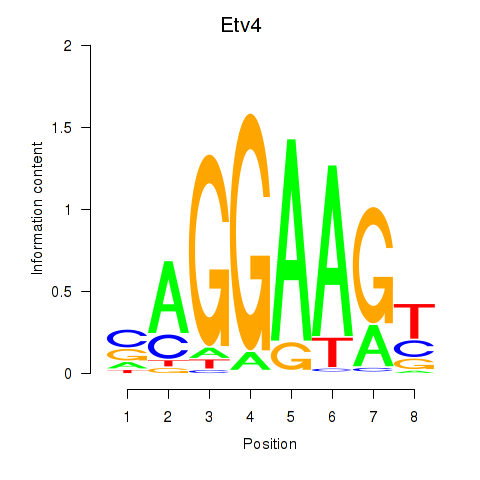
Activity profile of Etv4 motif
Sorted Z-values of Etv4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_158088505 | 18.96 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr4_+_163162211 | 15.88 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr5_+_82587420 | 14.50 |
ENSRNOT00000014020
|
Tlr4
|
toll-like receptor 4 |
| chr16_+_18716019 | 14.24 |
ENSRNOT00000047870
|
Sftpa1
|
surfactant protein A1 |
| chr10_+_83655460 | 12.38 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr10_+_47765432 | 12.35 |
ENSRNOT00000078231
|
LOC102553715
|
microfibril-associated glycoprotein 4-like |
| chr2_-_190100276 | 11.72 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr11_+_64601029 | 11.13 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr10_+_89167146 | 10.02 |
ENSRNOT00000043953
|
Ramp2
|
receptor activity modifying protein 2 |
| chr10_+_89166890 | 9.98 |
ENSRNOT00000088331
|
Ramp2
|
receptor activity modifying protein 2 |
| chr20_+_30915213 | 9.01 |
ENSRNOT00000000681
|
Prf1
|
perforin 1 |
| chr10_+_35133252 | 8.74 |
ENSRNOT00000051916
|
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr2_-_21698937 | 8.70 |
ENSRNOT00000080165
|
AABR07007642.1
|
|
| chr7_-_107392972 | 8.68 |
ENSRNOT00000093425
|
Tmem71
|
transmembrane protein 71 |
| chr3_+_55369214 | 8.65 |
ENSRNOT00000067161
|
Nostrin
|
nitric oxide synthase trafficking |
| chr13_+_47602692 | 8.58 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr1_-_98493978 | 8.44 |
ENSRNOT00000023942
|
Nkg7
|
natural killer cell granule protein 7 |
| chr13_-_55173692 | 8.22 |
ENSRNOT00000064785
ENSRNOT00000029878 ENSRNOT00000029865 ENSRNOT00000060292 ENSRNOT00000000814 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr7_+_142776252 | 7.87 |
ENSRNOT00000008673
|
Acvrl1
|
activin A receptor like type 1 |
| chr2_-_113616766 | 7.76 |
ENSRNOT00000016858
ENSRNOT00000074723 |
Tmem212
|
transmembrane protein 212 |
| chr16_-_19918644 | 7.38 |
ENSRNOT00000083345
ENSRNOT00000023926 |
Plvap
|
plasmalemma vesicle associated protein |
| chr7_+_142776580 | 7.36 |
ENSRNOT00000081047
|
Acvrl1
|
activin A receptor like type 1 |
| chr8_-_133002201 | 7.31 |
ENSRNOT00000008772
|
Ccr1
|
C-C motif chemokine receptor 1 |
| chr15_+_57221292 | 7.28 |
ENSRNOT00000014502
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr2_-_197860699 | 7.02 |
ENSRNOT00000028740
|
Ecm1
|
extracellular matrix protein 1 |
| chr3_-_160730360 | 6.81 |
ENSRNOT00000075864
|
RGD1563818
|
similar to secretory leukocyte protease inhibitor |
| chr2_-_172459165 | 6.76 |
ENSRNOT00000057473
|
Schip1
|
schwannomin interacting protein 1 |
| chr10_-_70646495 | 6.54 |
ENSRNOT00000071218
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr3_+_122114108 | 6.40 |
ENSRNOT00000091935
|
Sirpa
|
signal-regulatory protein alpha |
| chr18_+_51492196 | 6.14 |
ENSRNOT00000020570
|
Gramd3
|
GRAM domain containing 3 |
| chr5_-_141242131 | 5.74 |
ENSRNOT00000081482
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr17_-_46794845 | 5.70 |
ENSRNOT00000077910
ENSRNOT00000090663 ENSRNOT00000078331 |
Elmo1
|
engulfment and cell motility 1 |
| chr1_+_88875375 | 5.69 |
ENSRNOT00000028284
|
Tyrobp
|
Tyro protein tyrosine kinase binding protein |
| chr13_+_89774764 | 5.67 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr5_-_101138427 | 5.48 |
ENSRNOT00000058615
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr1_+_219403970 | 5.35 |
ENSRNOT00000029607
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr9_-_85986208 | 5.30 |
ENSRNOT00000073128
|
Fam124b
|
family with sequence similarity 124 member B |
| chr10_+_12046541 | 5.20 |
ENSRNOT00000081191
|
Mefv
|
MEFV, pyrin innate immunity regulator |
| chr5_-_137321121 | 5.18 |
ENSRNOT00000027414
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr15_-_54906203 | 5.04 |
ENSRNOT00000020186
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr3_-_110517163 | 5.01 |
ENSRNOT00000078037
|
Plcb2
|
phospholipase C, beta 2 |
| chr18_-_24735349 | 4.94 |
ENSRNOT00000036537
|
Gpr17
|
G protein-coupled receptor 17 |
| chr9_-_90806126 | 4.88 |
ENSRNOT00000072054
|
NEWGENE_1589866
|
family with sequence similarity 124B |
| chr5_-_160158386 | 4.74 |
ENSRNOT00000089345
|
Fblim1
|
filamin binding LIM protein 1 |
| chr13_+_50164563 | 4.71 |
ENSRNOT00000029533
|
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr7_+_143707237 | 4.67 |
ENSRNOT00000074212
|
Tns2
|
tensin 2 |
| chr1_+_198744050 | 4.65 |
ENSRNOT00000024404
|
Itgal
|
integrin subunit alpha L |
| chr8_-_72204730 | 4.62 |
ENSRNOT00000023810
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chrX_-_32355296 | 4.59 |
ENSRNOT00000081652
ENSRNOT00000065075 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr4_-_119327822 | 4.54 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr5_-_147303346 | 4.52 |
ENSRNOT00000009153
|
Hpca
|
hippocalcin |
| chr7_-_71048383 | 4.47 |
ENSRNOT00000005693
|
Gpr182
|
G protein-coupled receptor 182 |
| chr7_+_132857628 | 4.44 |
ENSRNOT00000005438
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr1_+_215628785 | 4.31 |
ENSRNOT00000054864
|
Lsp1
|
lymphocyte-specific protein 1 |
| chr10_+_14094754 | 4.27 |
ENSRNOT00000019660
|
Rpl3l
|
ribosomal protein L3-like |
| chr10_+_12046701 | 4.26 |
ENSRNOT00000011073
ENSRNOT00000084004 |
Mefv
|
MEFV, pyrin innate immunity regulator |
| chr13_+_51958834 | 4.25 |
ENSRNOT00000007833
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr1_-_227441442 | 4.23 |
ENSRNOT00000028433
|
Ms4a1
|
membrane spanning 4-domains A1 |
| chr10_-_30118873 | 4.19 |
ENSRNOT00000006063
|
Ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr10_+_91254058 | 4.14 |
ENSRNOT00000087218
ENSRNOT00000065373 |
Fmnl1
|
formin-like 1 |
| chr2_+_112868707 | 4.13 |
ENSRNOT00000017805
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr3_+_80555196 | 4.11 |
ENSRNOT00000067318
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr15_+_34352914 | 4.11 |
ENSRNOT00000067150
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr4_-_10329241 | 4.08 |
ENSRNOT00000017232
|
Fgl2
|
fibrinogen-like 2 |
| chrX_+_134979646 | 4.07 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr9_+_94745217 | 4.03 |
ENSRNOT00000051338
|
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr10_-_83655182 | 4.02 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr7_+_119482272 | 4.00 |
ENSRNOT00000009544
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr10_-_103685844 | 3.98 |
ENSRNOT00000064284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chr7_+_12782491 | 3.97 |
ENSRNOT00000065093
|
Cnn2
|
calponin 2 |
| chr10_-_64657089 | 3.90 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr11_-_66759402 | 3.90 |
ENSRNOT00000003326
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr3_+_79918969 | 3.77 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr9_+_42871950 | 3.76 |
ENSRNOT00000089673
|
Arid5a
|
AT-rich interaction domain 5A |
| chr1_+_88078350 | 3.72 |
ENSRNOT00000048677
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr2_-_243224883 | 3.70 |
ENSRNOT00000014139
|
Dapp1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides 1 |
| chr8_-_116465801 | 3.70 |
ENSRNOT00000085915
|
Sema3f
|
semaphorin 3F |
| chr10_+_63662986 | 3.69 |
ENSRNOT00000088722
|
Scarf1
|
scavenger receptor class F, member 1 |
| chr1_-_126211439 | 3.65 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr1_+_197999037 | 3.58 |
ENSRNOT00000091065
|
Apobr
|
apolipoprotein B receptor |
| chr10_-_64642292 | 3.55 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr16_-_9430743 | 3.53 |
ENSRNOT00000043811
|
Wdfy4
|
WDFY family member 4 |
| chr18_+_30550877 | 3.47 |
ENSRNOT00000027164
|
LOC108348201
|
protocadherin beta-7-like |
| chr14_+_91783514 | 3.44 |
ENSRNOT00000080753
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr8_-_65587658 | 3.43 |
ENSRNOT00000091982
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr3_+_11679530 | 3.41 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr1_-_199823386 | 3.40 |
ENSRNOT00000027375
|
Rgs10
|
regulator of G-protein signaling 10 |
| chr10_-_13892997 | 3.40 |
ENSRNOT00000004192
|
Traf7
|
TNF receptor associated factor 7 |
| chr8_-_65587427 | 3.34 |
ENSRNOT00000016491
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr1_+_60717386 | 3.30 |
ENSRNOT00000015019
|
Ppp2r1a
|
protein phosphatase 2 scaffold subunit A alpha |
| chr1_-_47502952 | 3.25 |
ENSRNOT00000025580
|
Tagap
|
T-cell activation RhoGTPase activating protein |
| chr14_-_83062302 | 3.15 |
ENSRNOT00000086769
ENSRNOT00000085735 |
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr7_+_145068286 | 3.14 |
ENSRNOT00000088956
ENSRNOT00000065753 |
Nckap1l
|
NCK associated protein 1 like |
| chr6_-_125317593 | 3.05 |
ENSRNOT00000074142
|
LOC102553138
|
ovarian cancer G-protein coupled receptor 1-like |
| chr14_-_86739335 | 3.02 |
ENSRNOT00000078282
|
Purb
|
purine rich element binding protein B |
| chr3_+_58965552 | 2.88 |
ENSRNOT00000002068
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr6_+_109939345 | 2.84 |
ENSRNOT00000013560
|
Ift43
|
intraflagellar transport 43 |
| chr12_-_39667849 | 2.79 |
ENSRNOT00000011499
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chrX_-_77700269 | 2.77 |
ENSRNOT00000092418
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr1_+_256786124 | 2.75 |
ENSRNOT00000034563
|
Ffar4
|
free fatty acid receptor 4 |
| chr10_-_56270640 | 2.70 |
ENSRNOT00000056918
|
Cd68
|
Cd68 molecule |
| chr6_+_98284170 | 2.67 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr5_-_59063592 | 2.60 |
ENSRNOT00000022400
|
Tln1
|
talin 1 |
| chrX_-_71616997 | 2.58 |
ENSRNOT00000004406
|
Cxcr3
|
C-X-C motif chemokine receptor 3 |
| chr20_+_27954433 | 2.52 |
ENSRNOT00000064288
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr1_+_198528635 | 2.51 |
ENSRNOT00000022765
|
LOC308990
|
hypothetical protein LOC308990 |
| chr20_-_5441706 | 2.41 |
ENSRNOT00000000549
|
Vps52
|
VPS52 GARP complex subunit |
| chr15_-_34469350 | 2.41 |
ENSRNOT00000067536
|
Adcy4
|
adenylate cyclase 4 |
| chr20_-_4818568 | 2.41 |
ENSRNOT00000001115
|
Ddx39b
|
DExD-box helicase 39B |
| chr8_-_118893702 | 2.40 |
ENSRNOT00000028416
|
Nradd
|
neurotrophin receptor associated death domain |
| chr9_+_94879745 | 2.39 |
ENSRNOT00000080482
|
Atg16l1
|
autophagy related 16-like 1 |
| chr2_+_243160917 | 2.38 |
ENSRNOT00000014027
|
Lamtor3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr5_+_144031402 | 2.34 |
ENSRNOT00000011694
|
Csf3r
|
colony stimulating factor 3 receptor |
| chr10_-_90995982 | 2.33 |
ENSRNOT00000093266
|
Gfap
|
glial fibrillary acidic protein |
| chr8_-_82641536 | 2.32 |
ENSRNOT00000014491
|
Scg3
|
secretogranin III |
| chr15_-_50425900 | 2.31 |
ENSRNOT00000058715
|
Adam28
|
ADAM metallopeptidase domain 28 |
| chr3_-_164239250 | 2.28 |
ENSRNOT00000012604
|
Spata2
|
spermatogenesis associated 2 |
| chr3_+_28627084 | 2.27 |
ENSRNOT00000049884
|
Arhgap15
|
Rho GTPase activating protein 15 |
| chr14_-_86333424 | 2.25 |
ENSRNOT00000083191
|
Nudcd3
|
NudC domain containing 3 |
| chr20_+_4355175 | 2.24 |
ENSRNOT00000000510
|
Gpsm3
|
G-protein signaling modulator 3 |
| chr6_+_43001948 | 2.21 |
ENSRNOT00000007374
|
Hpcal1
|
hippocalcin-like 1 |
| chr15_-_57805184 | 2.20 |
ENSRNOT00000000168
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr11_-_60882379 | 2.17 |
ENSRNOT00000002799
|
Cd200r1
|
CD200 receptor 1 |
| chr5_-_140585408 | 2.12 |
ENSRNOT00000018711
|
Cap1
|
adenylate cyclase associated protein 1 |
| chr3_+_161272385 | 2.12 |
ENSRNOT00000021052
|
Zswim3
|
zinc finger, SWIM-type containing 3 |
| chr8_+_117062884 | 2.10 |
ENSRNOT00000082452
ENSRNOT00000071540 |
Nicn1
|
nicolin 1 |
| chr20_+_5455974 | 2.09 |
ENSRNOT00000000553
ENSRNOT00000092676 |
Pfdn6
|
prefoldin subunit 6 |
| chr10_-_14299167 | 2.06 |
ENSRNOT00000042066
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr3_-_80875817 | 2.06 |
ENSRNOT00000091265
|
Dgkz
|
diacylglycerol kinase zeta |
| chr1_+_142050458 | 2.02 |
ENSRNOT00000018107
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr13_-_88536728 | 2.00 |
ENSRNOT00000003950
|
Uhmk1
|
U2AF homology motif kinase 1 |
| chr18_+_29629184 | 1.98 |
ENSRNOT00000021797
|
Hars2
|
histidyl-tRNA synthetase 2 |
| chr16_-_7588841 | 1.98 |
ENSRNOT00000084645
|
Mettl6
|
methyltransferase like 6 |
| chr1_+_197999336 | 1.95 |
ENSRNOT00000023555
|
Apobr
|
apolipoprotein B receptor |
| chr18_+_56379890 | 1.95 |
ENSRNOT00000078764
|
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr1_-_221015929 | 1.94 |
ENSRNOT00000028137
|
Sipa1
|
signal-induced proliferation-associated 1 |
| chr8_-_122604787 | 1.92 |
ENSRNOT00000047198
|
Trim71
|
tripartite motif containing 71 |
| chr5_+_144160108 | 1.91 |
ENSRNOT00000064972
|
Eva1b
|
eva-1 homolog B |
| chr1_+_8310577 | 1.90 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr7_-_11223649 | 1.87 |
ENSRNOT00000061191
|
Mfsd12
|
major facilitator superfamily domain containing 12 |
| chr1_+_142060955 | 1.83 |
ENSRNOT00000017862
|
Vps33b
|
VPS33B, late endosome and lysosome associated |
| chr12_+_43940929 | 1.81 |
ENSRNOT00000001486
|
Rnft2
|
ring finger protein, transmembrane 2 |
| chr18_+_51523758 | 1.79 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr5_+_153507093 | 1.79 |
ENSRNOT00000086650
ENSRNOT00000083645 |
Runx3
|
runt-related transcription factor 3 |
| chr11_-_65845418 | 1.76 |
ENSRNOT00000091057
|
Fstl1
|
follistatin-like 1 |
| chrX_-_78911601 | 1.75 |
ENSRNOT00000003188
|
RGD1566265
|
similar to RIKEN cDNA 2610002M06 |
| chr1_-_77844189 | 1.66 |
ENSRNOT00000017555
|
Gltscr2
|
glioma tumor suppressor candidate region gene 2 |
| chr1_-_198662610 | 1.65 |
ENSRNOT00000055012
|
Sept1
|
septin 1 |
| chr3_-_111037166 | 1.64 |
ENSRNOT00000017070
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr16_-_19308842 | 1.63 |
ENSRNOT00000019556
|
Fam32a
|
family with sequence similarity 32, member A |
| chr5_+_64294321 | 1.62 |
ENSRNOT00000083796
|
Msantd3
|
Myb/SANT DNA binding domain containing 3 |
| chr4_+_6559545 | 1.61 |
ENSRNOT00000067183
|
Prkag2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr2_+_202470487 | 1.58 |
ENSRNOT00000026953
|
Gdap2
|
ganglioside-induced differentiation-associated-protein 2 |
| chr14_+_106008086 | 1.58 |
ENSRNOT00000009319
|
Peli1
|
pellino E3 ubiquitin protein ligase 1 |
| chr1_+_101055622 | 1.58 |
ENSRNOT00000079189
|
Nosip
|
nitric oxide synthase interacting protein |
| chr20_+_11168298 | 1.57 |
ENSRNOT00000032240
|
Trappc10
|
trafficking protein particle complex 10 |
| chr11_-_14304603 | 1.55 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr13_+_89797800 | 1.55 |
ENSRNOT00000005811
|
Usf1
|
upstream transcription factor 1 |
| chr2_-_187133993 | 1.52 |
ENSRNOT00000019502
|
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr13_-_95250235 | 1.47 |
ENSRNOT00000085648
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr3_-_162581610 | 1.45 |
ENSRNOT00000079324
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr3_+_67849966 | 1.44 |
ENSRNOT00000057826
|
Dusp19
|
dual specificity phosphatase 19 |
| chrX_-_15627235 | 1.42 |
ENSRNOT00000013369
|
Wdr45
|
WD repeat domain 45 |
| chr17_+_9837402 | 1.41 |
ENSRNOT00000076436
|
Rab24
|
RAB24, member RAS oncogene family |
| chr15_-_34479741 | 1.41 |
ENSRNOT00000027759
|
Ripk3
|
receptor-interacting serine-threonine kinase 3 |
| chr3_+_122544788 | 1.40 |
ENSRNOT00000063828
|
Tgm3
|
transglutaminase 3 |
| chr8_-_127782070 | 1.40 |
ENSRNOT00000045493
|
Plcd1
|
phospholipase C, delta 1 |
| chr5_-_137238354 | 1.40 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr3_+_171037957 | 1.40 |
ENSRNOT00000008764
|
Rbm38
|
RNA binding motif protein 38 |
| chr13_+_90467265 | 1.39 |
ENSRNOT00000008518
|
Copa
|
coatomer protein complex subunit alpha |
| chr1_-_20155960 | 1.38 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr3_+_160467552 | 1.38 |
ENSRNOT00000066657
|
Stk4
|
serine/threonine kinase 4 |
| chrY_+_914045 | 1.36 |
ENSRNOT00000088593
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr15_-_33766438 | 1.36 |
ENSRNOT00000033977
|
Ap1g2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chr18_-_58423196 | 1.36 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr10_+_55492404 | 1.33 |
ENSRNOT00000005588
ENSRNOT00000078038 |
Rpl26
|
ribosomal protein L26 |
| chr6_-_76552559 | 1.33 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr1_+_31576183 | 1.31 |
ENSRNOT00000019735
|
Pdcd6
|
programmed cell death 6 |
| chrX_+_15155230 | 1.31 |
ENSRNOT00000073289
ENSRNOT00000051439 |
Was
|
Wiskott-Aldrich syndrome |
| chr7_+_127964752 | 1.31 |
ENSRNOT00000038168
|
RGD1560617
|
hypothetical gene supported by NM_053561; AF062594 |
| chr8_-_103298927 | 1.30 |
ENSRNOT00000046873
ENSRNOT00000011609 |
U2surp
|
U2 snRNP-associated SURP domain containing |
| chr1_+_87019975 | 1.28 |
ENSRNOT00000041205
|
Lgals4
|
galectin 4 |
| chr13_+_90943255 | 1.28 |
ENSRNOT00000011539
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr8_-_21968415 | 1.27 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr10_-_84976170 | 1.25 |
ENSRNOT00000013542
|
Lrrc46
|
leucine rich repeat containing 46 |
| chr18_+_22964210 | 1.25 |
ENSRNOT00000066816
|
Pik3c3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr3_+_72080630 | 1.23 |
ENSRNOT00000008911
|
Med19
|
mediator complex subunit 19 |
| chr3_-_113423473 | 1.22 |
ENSRNOT00000064982
|
Serinc4
|
serine incorporator 4 |
| chr10_-_94988461 | 1.19 |
ENSRNOT00000048490
|
Ddx5
|
DEAD-box helicase 5 |
| chr5_-_60658521 | 1.18 |
ENSRNOT00000092866
ENSRNOT00000093024 ENSRNOT00000016875 |
Tomm5
Fbxo10
|
translocase of outer mitochondrial membrane 5 F-box protein 10 |
| chr15_+_34270648 | 1.17 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr15_-_19575914 | 1.15 |
ENSRNOT00000043897
|
AABR07017253.1
|
|
| chr7_-_93826665 | 1.14 |
ENSRNOT00000011344
|
Tnfrsf11b
|
TNF receptor superfamily member 11B |
| chr13_-_90467235 | 1.13 |
ENSRNOT00000081534
ENSRNOT00000008150 |
Ncstn
|
nicastrin |
| chr1_-_198316882 | 1.11 |
ENSRNOT00000085304
ENSRNOT00000064985 |
Taok2
|
TAO kinase 2 |
| chr3_-_111037425 | 1.09 |
ENSRNOT00000085628
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr17_-_21705773 | 1.09 |
ENSRNOT00000078010
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr5_-_154319629 | 1.09 |
ENSRNOT00000014415
|
Lypla2
|
lysophospholipase II |
| chr4_-_27473150 | 1.07 |
ENSRNOT00000032505
|
Krit1
|
KRIT1, ankyrin repeat containing |
| chr1_+_234749568 | 1.07 |
ENSRNOT00000016871
|
Ostf1
|
osteoclast stimulating factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Etv4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 3.6 | 14.5 | GO:0070428 | detection of lipopolysaccharide(GO:0032497) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 2.7 | 8.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 2.5 | 7.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 2.4 | 9.5 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 2.3 | 23.4 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 2.1 | 19.0 | GO:0070417 | cellular response to cold(GO:0070417) |
| 2.0 | 4.0 | GO:2000426 | negative regulation of apoptotic cell clearance(GO:2000426) |
| 1.9 | 15.2 | GO:0090500 | dorsal aorta morphogenesis(GO:0035912) endocardial cushion to mesenchymal transition(GO:0090500) |
| 1.9 | 7.5 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 1.8 | 1.8 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 1.6 | 14.2 | GO:0008228 | opsonization(GO:0008228) |
| 1.5 | 4.5 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) |
| 1.5 | 4.4 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) regulation of peroxidase activity(GO:2000468) |
| 1.3 | 1.3 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 1.3 | 7.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 1.0 | 3.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.0 | 3.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.0 | 3.9 | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.9 | 3.8 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.8 | 8.6 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.7 | 5.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.7 | 4.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.7 | 3.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.7 | 1.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.7 | 2.0 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.6 | 1.9 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.6 | 15.9 | GO:0030220 | platelet formation(GO:0030220) |
| 0.6 | 5.7 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.6 | 14.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.6 | 3.7 | GO:0097491 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.6 | 7.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.6 | 10.0 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.6 | 1.7 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.5 | 1.5 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) positive regulation of transcription by glucose(GO:0046016) |
| 0.5 | 1.4 | GO:2000449 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.5 | 4.7 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.5 | 2.3 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.5 | 8.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.5 | 3.6 | GO:0071000 | response to magnetism(GO:0071000) |
| 0.4 | 2.4 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.4 | 1.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.4 | 4.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.4 | 2.9 | GO:0033622 | integrin activation(GO:0033622) |
| 0.4 | 2.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.4 | 1.4 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 0.3 | 1.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 6.8 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.3 | 1.3 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.3 | 5.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.3 | 7.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.3 | 5.8 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.3 | 4.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 1.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 4.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.3 | 2.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.3 | 3.7 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.3 | 4.2 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.3 | 1.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.3 | 2.4 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.3 | 2.4 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.3 | 2.7 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.3 | 1.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.3 | 1.6 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.3 | 1.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.3 | 5.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 3.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 6.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 1.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.2 | 1.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 2.6 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.2 | 1.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 2.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 3.4 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.2 | 5.0 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.2 | 0.4 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 0.6 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) |
| 0.2 | 0.8 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 1.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 0.7 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.2 | 1.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 4.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.2 | 5.7 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.2 | 1.0 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.2 | 1.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.2 | 1.9 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 6.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 2.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 3.5 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.2 | 0.5 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 6.6 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.2 | 4.1 | GO:0007567 | parturition(GO:0007567) |
| 0.2 | 0.5 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 4.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 5.2 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 2.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.4 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.1 | 1.6 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 0.9 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 5.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.5 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 2.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 1.7 | GO:1901673 | cell separation after cytokinesis(GO:0000920) regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 2.0 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.9 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 3.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 1.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.9 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 1.8 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 1.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.3 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 4.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.6 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.2 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.1 | 4.1 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 4.3 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 0.7 | GO:0051934 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) |
| 0.1 | 0.6 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.6 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.1 | 1.6 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.7 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 5.5 | GO:0006641 | triglyceride metabolic process(GO:0006641) |
| 0.1 | 0.6 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 2.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 1.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 1.5 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.0 | 1.2 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 19.1 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 2.1 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.5 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 1.7 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 2.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.4 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 1.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.7 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 2.1 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.5 | GO:0002687 | positive regulation of leukocyte migration(GO:0002687) |
| 0.0 | 0.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 2.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 1.8 | GO:0051607 | defense response to virus(GO:0051607) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 19.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 2.9 | 14.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 2.2 | 4.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.7 | 5.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.7 | 5.0 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 1.5 | 9.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.6 | 2.4 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.6 | 2.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.5 | 4.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.4 | 6.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 2.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 4.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 14.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.4 | 4.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 3.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 2.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.3 | 1.0 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.3 | 2.0 | GO:0089701 | U2AF(GO:0089701) |
| 0.3 | 8.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 2.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 3.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 2.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.3 | 2.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.3 | 2.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 2.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 9.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 1.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.2 | 1.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 3.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 1.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 0.8 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 1.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 15.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 1.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 1.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 13.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 3.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.6 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 1.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 2.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 7.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.4 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 1.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 1.0 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 18.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 35.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 2.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 6.3 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 7.4 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.4 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 2.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 1.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.6 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 1.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 5.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 30.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 3.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.1 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 2.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 6.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 5.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 3.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.5 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 2.4 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 6.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 19.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.5 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 2.6 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 6.4 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 1.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 2.5 | 15.2 | GO:0098821 | activin receptor activity, type I(GO:0016361) BMP receptor activity(GO:0098821) |
| 2.4 | 7.3 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 2.0 | 11.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.7 | 5.0 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 1.6 | 7.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 1.5 | 4.5 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 1.5 | 4.4 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 1.4 | 7.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.3 | 6.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.1 | 20.0 | GO:0015026 | coreceptor activity(GO:0015026) |
| 1.1 | 5.5 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 1.0 | 4.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) PTB domain binding(GO:0051425) |
| 1.0 | 3.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.0 | 4.0 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.9 | 8.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.8 | 2.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.7 | 19.0 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.7 | 3.4 | GO:0005534 | galactose binding(GO:0005534) |
| 0.7 | 2.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.5 | 1.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.5 | 1.5 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.5 | 3.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.5 | 1.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.5 | 3.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 4.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.4 | 2.6 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.4 | 4.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 1.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 4.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.3 | 2.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 6.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 2.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.3 | 2.1 | GO:0017070 | U6 snRNA binding(GO:0017070) U4 snRNA binding(GO:0030621) |
| 0.3 | 3.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 1.0 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.3 | 1.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.3 | 9.0 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.2 | 4.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 5.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.2 | 29.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 4.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 3.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 0.6 | GO:0019961 | interferon binding(GO:0019961) |
| 0.2 | 0.6 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.2 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 0.7 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.2 | 2.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.2 | 3.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 1.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 1.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 4.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 4.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 2.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 34.3 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 7.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 2.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 1.2 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.1 | 1.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 2.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 27.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 23.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 0.5 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 1.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 1.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.4 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 4.5 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 2.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 7.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 2.3 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 1.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 1.0 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 40.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.5 | 18.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.4 | 12.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.4 | 9.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.4 | 14.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.4 | 4.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 4.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.3 | 7.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 3.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 11.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.3 | 7.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 11.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 4.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 8.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.2 | 3.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 25.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 3.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 2.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 4.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 22.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 4.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 0.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 14.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 1.3 | 19.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.9 | 12.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.8 | 9.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.8 | 17.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.8 | 12.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.7 | 7.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.6 | 4.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.5 | 10.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.5 | 7.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 5.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 5.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 3.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 3.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 3.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.3 | 20.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 36.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 1.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 1.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.2 | 9.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 1.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 6.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 2.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 1.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.0 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.9 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 2.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |