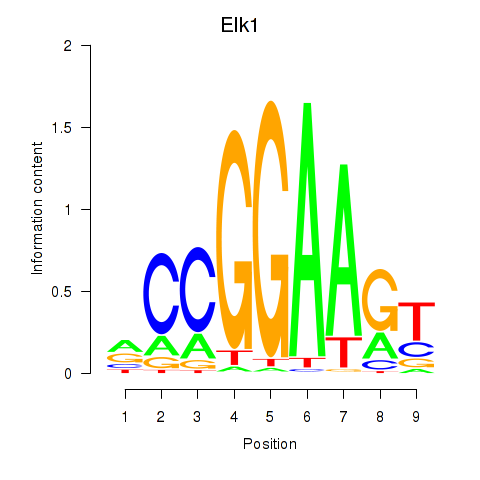
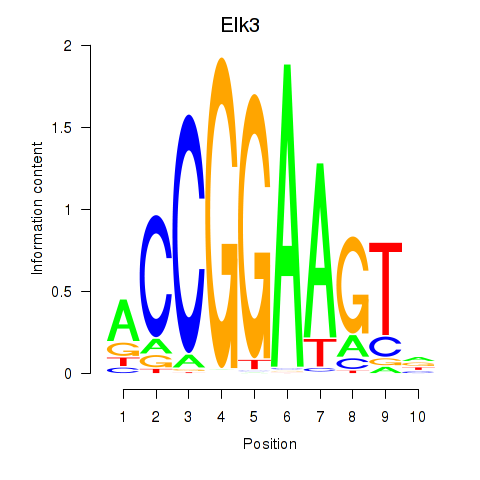
Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
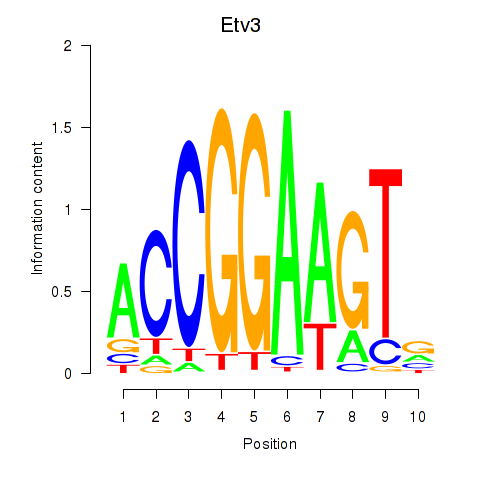
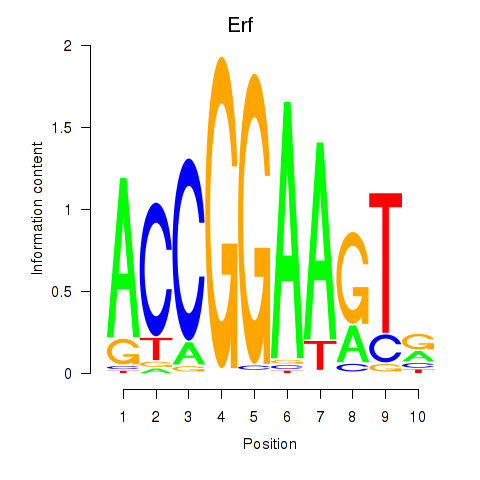
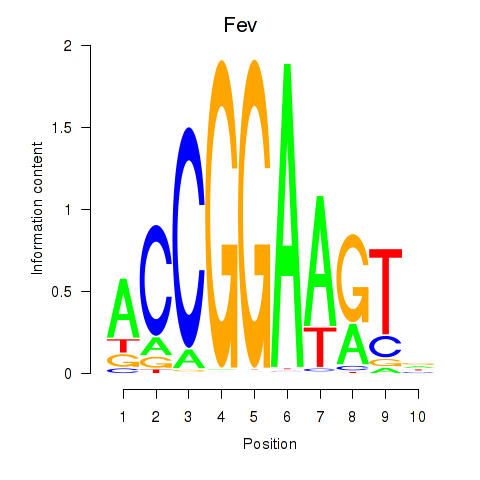
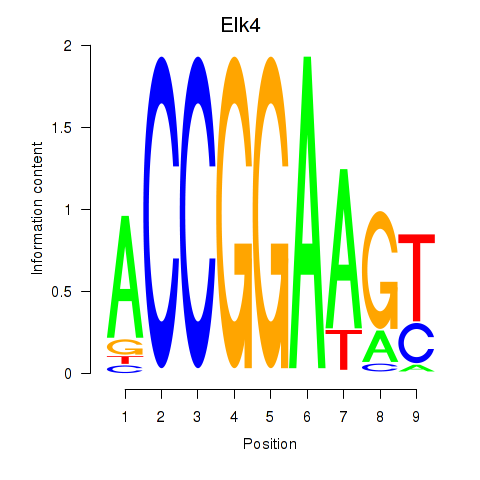
Results for Etv3_Erf_Fev_Elk4_Elk1_Elk3
Z-value: 2.64
Transcription factors associated with Etv3_Erf_Fev_Elk4_Elk1_Elk3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv3
|
ENSRNOG00000043095 | ets variant 3 |
|
Erf
|
ENSRNOG00000020426 | Ets2 repressor factor |
|
Fev
|
ENSRNOG00000017856 | FEV, ETS transcription factor |
|
Elk4
|
ENSRNOG00000007887 | ELK4, ETS transcription factor |
|
Elk1
|
ENSRNOG00000010171 | ELK1, ETS transcription factor |
|
Elk3
|
ENSRNOG00000004367 | ELK3, ETS-domain protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Elk4 | rn6_v1_chr13_+_48790951_48790951 | -0.85 | 1.3e-89 | Click! |
| Etv3 | rn6_v1_chr2_+_186872520_186872572 | -0.78 | 7.4e-66 | Click! |
| Elk3 | rn6_v1_chr7_-_34121694_34121694 | -0.52 | 1.5e-23 | Click! |
| Elk1 | rn6_v1_chrX_+_1297099_1297099 | 0.32 | 3.4e-09 | Click! |
| Fev | rn6_v1_chr9_-_82146874_82146874 | 0.12 | 3.2e-02 | Click! |
| Erf | rn6_v1_chr1_-_82120902_82120902 | -0.02 | 6.7e-01 | Click! |
Activity profile of Etv3_Erf_Fev_Elk4_Elk1_Elk3 motif
Sorted Z-values of Etv3_Erf_Fev_Elk4_Elk1_Elk3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_34166599 | 260.79 |
ENSRNOT00000003246
|
Trim41
|
tripartite motif-containing 41 |
| chr8_-_65587427 | 159.49 |
ENSRNOT00000016491
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr13_-_37287458 | 142.63 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr8_-_65587658 | 134.50 |
ENSRNOT00000091982
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr20_+_5125349 | 104.95 |
ENSRNOT00000085598
ENSRNOT00000001129 |
Bag6
|
BCL2-associated athanogene 6 |
| chr6_+_104291071 | 99.00 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr15_-_34352673 | 93.98 |
ENSRNOT00000064916
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr2_+_115337439 | 90.83 |
ENSRNOT00000015779
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr6_-_104290579 | 87.48 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr16_+_21275311 | 85.25 |
ENSRNOT00000027980
|
LOC100911483
|
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-like |
| chr3_-_168018410 | 84.49 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr2_-_18927365 | 83.18 |
ENSRNOT00000045850
|
Xrcc4
|
X-ray repair cross complementing 4 |
| chr20_+_5455974 | 82.63 |
ENSRNOT00000000553
ENSRNOT00000092676 |
Pfdn6
|
prefoldin subunit 6 |
| chr10_-_27366665 | 81.83 |
ENSRNOT00000004725
|
Gabra1
|
gamma-aminobutyric acid type A receptor alpha1 subunit |
| chr19_+_56010085 | 79.62 |
ENSRNOT00000058216
|
Spata33
|
spermatogenesis associated 33 |
| chr20_+_6869767 | 79.15 |
ENSRNOT00000000631
ENSRNOT00000086330 ENSRNOT00000093188 ENSRNOT00000093736 |
RGD735065
|
similar to GI:13385412-like protein splice form I |
| chr3_+_60024013 | 78.74 |
ENSRNOT00000025255
|
Scrn3
|
secernin 3 |
| chr10_+_14094754 | 78.32 |
ENSRNOT00000019660
|
Rpl3l
|
ribosomal protein L3-like |
| chr7_-_119158173 | 75.61 |
ENSRNOT00000067483
ENSRNOT00000078528 |
Txn2
|
thioredoxin 2 |
| chr3_+_80555196 | 75.28 |
ENSRNOT00000067318
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr15_-_4057104 | 75.23 |
ENSRNOT00000084350
ENSRNOT00000012270 |
Sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr16_+_71058022 | 72.74 |
ENSRNOT00000066901
|
Bag4
|
BCL2-associated athanogene 4 |
| chr1_+_225120061 | 71.35 |
ENSRNOT00000026875
|
B3gat3
|
beta-1,3-glucuronyltransferase 3 |
| chr20_-_5441706 | 66.53 |
ENSRNOT00000000549
|
Vps52
|
VPS52 GARP complex subunit |
| chr19_-_11451278 | 64.72 |
ENSRNOT00000026118
|
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr20_+_5125679 | 64.36 |
ENSRNOT00000060832
|
Bag6
|
BCL2-associated athanogene 6 |
| chr5_+_142702685 | 63.25 |
ENSRNOT00000085986
ENSRNOT00000010373 ENSRNOT00000087416 |
Sf3a3
|
splicing factor 3a, subunit 3 |
| chr6_-_108076186 | 63.22 |
ENSRNOT00000014814
|
Fam161b
|
family with sequence similarity 161, member B |
| chr11_+_64790801 | 62.39 |
ENSRNOT00000004023
|
Timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chrX_+_26294066 | 62.09 |
ENSRNOT00000037862
|
Hccs
|
holocytochrome c synthase |
| chr9_-_23352668 | 61.05 |
ENSRNOT00000075279
|
Mut
|
methylmalonyl CoA mutase |
| chr13_-_89668473 | 61.05 |
ENSRNOT00000004938
|
Ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr8_+_70522092 | 60.69 |
ENSRNOT00000025873
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr3_-_151688149 | 60.40 |
ENSRNOT00000072945
|
Nfs1
|
NFS1 cysteine desulfurase |
| chr17_-_79798435 | 60.10 |
ENSRNOT00000022986
|
Fam188a
|
family with sequence similarity 188, member A |
| chr11_-_27080701 | 60.05 |
ENSRNOT00000002180
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr4_+_27647335 | 59.64 |
ENSRNOT00000067376
|
Gatad1
|
GATA zinc finger domain containing 1 |
| chr6_+_108936664 | 58.45 |
ENSRNOT00000007298
|
Dlst
|
dihydrolipoamide S-succinyltransferase |
| chr15_+_12407524 | 58.18 |
ENSRNOT00000009249
|
Psmd6
|
proteasome 26S subunit, non-ATPase 6 |
| chr19_+_37282018 | 57.79 |
ENSRNOT00000021723
|
Tmem208
|
transmembrane protein 208 |
| chr4_-_9881484 | 57.51 |
ENSRNOT00000016450
|
Psmc2
|
proteasome 26S subunit, ATPase 2 |
| chr6_-_64170122 | 57.34 |
ENSRNOT00000093248
ENSRNOT00000005363 |
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr3_-_79728879 | 56.98 |
ENSRNOT00000012425
|
Ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3 |
| chr15_-_57805184 | 56.79 |
ENSRNOT00000000168
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr6_-_8346197 | 56.58 |
ENSRNOT00000061826
|
Prepl
|
prolyl endopeptidase-like |
| chr10_-_74001895 | 54.83 |
ENSRNOT00000005658
|
Vmp1
|
vacuole membrane protein 1 |
| chr3_-_152222647 | 53.30 |
ENSRNOT00000026820
|
Nfs1
|
NFS1 cysteine desulfurase |
| chr9_+_94879745 | 53.29 |
ENSRNOT00000080482
|
Atg16l1
|
autophagy related 16-like 1 |
| chr4_+_118655728 | 52.98 |
ENSRNOT00000043082
|
Aak1
|
AP2 associated kinase 1 |
| chr17_+_15966234 | 52.84 |
ENSRNOT00000084304
|
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr2_+_244521699 | 51.25 |
ENSRNOT00000029382
|
Stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr1_+_142050458 | 50.84 |
ENSRNOT00000018107
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr6_-_22138286 | 50.76 |
ENSRNOT00000007607
|
Yipf4
|
Yip1 domain family, member 4 |
| chr8_+_126411829 | 50.72 |
ENSRNOT00000013450
|
Azi2
|
5-azacytidine induced 2 |
| chr8_+_21663325 | 49.78 |
ENSRNOT00000027749
|
Ubl5
|
ubiquitin-like 5 |
| chr7_-_36408588 | 49.54 |
ENSRNOT00000063946
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr1_-_48033343 | 48.58 |
ENSRNOT00000019531
ENSRNOT00000076422 |
Tcp1
|
t-complex 1 |
| chr14_-_86333424 | 48.28 |
ENSRNOT00000083191
|
Nudcd3
|
NudC domain containing 3 |
| chr7_+_26256459 | 47.64 |
ENSRNOT00000010986
|
Appl2
|
adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 2 |
| chr8_+_49354115 | 47.60 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr3_+_151688454 | 47.43 |
ENSRNOT00000026856
|
Romo1
|
reactive oxygen species modulator 1 |
| chr3_+_152222726 | 47.13 |
ENSRNOT00000072464
|
LOC100910944
|
reactive oxygen species modulator 1-like |
| chr20_-_45005035 | 46.94 |
ENSRNOT00000035047
|
Mfsd4b
|
major facilitator superfamily domain containing 4B |
| chr9_+_10846596 | 46.41 |
ENSRNOT00000075386
|
Dpp9
|
dipeptidyl peptidase 9 |
| chr7_-_12918173 | 46.25 |
ENSRNOT00000011010
|
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr15_-_58711872 | 45.95 |
ENSRNOT00000058204
|
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr9_-_99818262 | 45.87 |
ENSRNOT00000056600
|
Cops9
|
COP9 signalosome subunit 9 |
| chr19_+_22281906 | 45.71 |
ENSRNOT00000021711
|
Itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr1_+_31576183 | 45.48 |
ENSRNOT00000019735
|
Pdcd6
|
programmed cell death 6 |
| chr11_+_46179940 | 45.23 |
ENSRNOT00000088152
|
Tfg
|
Trk-fused gene |
| chr16_-_2367253 | 44.53 |
ENSRNOT00000083573
|
Pde12
|
phosphodiesterase 12 |
| chr9_-_69953182 | 44.13 |
ENSRNOT00000015852
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr15_+_34352914 | 43.85 |
ENSRNOT00000067150
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr1_-_89084859 | 43.67 |
ENSRNOT00000032026
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr11_+_83868655 | 43.54 |
ENSRNOT00000072402
|
NEWGENE_621438
|
thrombopoietin |
| chr13_-_82607379 | 43.16 |
ENSRNOT00000051763
|
Blzf1
|
basic leucine zipper nuclear factor 1 |
| chr10_-_46206135 | 42.54 |
ENSRNOT00000091471
|
Cops3
|
COP9 signalosome subunit 3 |
| chr3_-_122813583 | 42.49 |
ENSRNOT00000009681
|
Idh3B
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr11_+_16826399 | 42.33 |
ENSRNOT00000050701
|
Cxadr
|
coxsackie virus and adenovirus receptor |
| chr4_-_56897310 | 41.87 |
ENSRNOT00000043902
ENSRNOT00000090038 |
Tnpo3
|
transportin 3 |
| chr6_+_34094306 | 41.73 |
ENSRNOT00000051970
|
Wdr35
|
WD repeat domain 35 |
| chr19_+_551708 | 41.52 |
ENSRNOT00000015811
|
Fam96b
|
family with sequence similarity 96, member B |
| chr1_+_192088672 | 41.51 |
ENSRNOT00000024367
|
Dctn5
|
dynactin subunit 5 |
| chr1_-_165382216 | 41.36 |
ENSRNOT00000023648
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr13_-_101697684 | 41.32 |
ENSRNOT00000078834
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr3_-_164986180 | 41.23 |
ENSRNOT00000016150
|
Dpm1
|
dolichyl-phosphate mannosyltransferase subunit 1, catalytic |
| chr2_+_218951451 | 41.15 |
ENSRNOT00000019190
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr10_-_86355006 | 40.91 |
ENSRNOT00000075076
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr3_+_140106766 | 40.43 |
ENSRNOT00000014046
|
Naa20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr13_-_97838228 | 40.43 |
ENSRNOT00000003618
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr17_+_47397558 | 40.07 |
ENSRNOT00000085923
|
Epdr1
|
ependymin related 1 |
| chr20_-_38985036 | 39.71 |
ENSRNOT00000001066
|
Serinc1
|
serine incorporator 1 |
| chr3_-_22951711 | 39.40 |
ENSRNOT00000016876
ENSRNOT00000085981 |
Psmb7
|
proteasome subunit beta 7 |
| chr7_+_98813040 | 39.32 |
ENSRNOT00000012616
|
Ndufb9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr3_+_94848823 | 38.99 |
ENSRNOT00000041362
|
Ccdc73
|
coiled-coil domain containing 73 |
| chr17_+_5225835 | 38.94 |
ENSRNOT00000022373
|
Zcchc6
|
zinc finger CCHC-type containing 6 |
| chr14_+_2325308 | 38.91 |
ENSRNOT00000000072
|
Atp5i
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr19_+_52032886 | 38.32 |
ENSRNOT00000019923
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr12_+_19231092 | 38.30 |
ENSRNOT00000045379
|
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr1_-_261140389 | 37.97 |
ENSRNOT00000073193
|
Rrp12
|
ribosomal RNA processing 12 homolog |
| chr1_-_174620064 | 37.78 |
ENSRNOT00000016224
|
Tmem41b
|
transmembrane protein 41B |
| chr19_-_22281778 | 37.74 |
ENSRNOT00000049624
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr10_+_82375572 | 37.34 |
ENSRNOT00000004947
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr16_-_71057883 | 37.15 |
ENSRNOT00000020721
|
Lsm1
|
LSM1 homolog, mRNA degradation associated |
| chr10_+_72197977 | 37.02 |
ENSRNOT00000003886
|
Myo19
|
myosin XIX |
| chr8_+_22625874 | 36.71 |
ENSRNOT00000012269
|
Timm29
|
translocase of inner mitochondrial membrane 29 |
| chr2_-_183582553 | 36.67 |
ENSRNOT00000014236
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr2_+_4195917 | 36.58 |
ENSRNOT00000093303
|
RGD1560883
|
similar to KIAA0825 protein |
| chr4_+_56625561 | 36.28 |
ENSRNOT00000008356
ENSRNOT00000090808 ENSRNOT00000008972 |
Calu
|
calumenin |
| chr1_+_247110216 | 35.66 |
ENSRNOT00000016000
|
Cdc37l1
|
cell division cycle 37-like 1 |
| chr3_-_92242318 | 35.61 |
ENSRNOT00000007018
|
Trim44
|
tripartite motif-containing 44 |
| chr7_-_117140168 | 35.55 |
ENSRNOT00000056481
ENSRNOT00000013353 |
Puf60
|
poly-U binding splicing factor 60 |
| chr1_+_189665976 | 35.49 |
ENSRNOT00000033673
|
Lyrm1
|
LYR motif containing 1 |
| chr8_+_113603533 | 35.37 |
ENSRNOT00000017280
|
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chr4_+_155709613 | 35.21 |
ENSRNOT00000013272
|
Necap1
|
NECAP endocytosis associated 1 |
| chr5_-_127523089 | 35.19 |
ENSRNOT00000016954
|
Cpt2
|
carnitine palmitoyltransferase 2 |
| chr7_-_95310005 | 35.08 |
ENSRNOT00000005815
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr4_-_157486844 | 34.80 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr3_-_60105212 | 34.68 |
ENSRNOT00000086329
ENSRNOT00000025158 |
Gpr155
|
G protein-coupled receptor 155 |
| chr5_+_104941066 | 34.67 |
ENSRNOT00000009225
|
Rraga
|
Ras-related GTP binding A |
| chr5_-_135397591 | 34.65 |
ENSRNOT00000050434
|
Tmem69
|
transmembrane protein 69 |
| chr8_-_50199978 | 34.45 |
ENSRNOT00000023459
|
Rnf214
|
ring finger protein 214 |
| chr11_-_45510961 | 34.44 |
ENSRNOT00000002238
|
Tomm70
|
translocase of outer mitochondrial membrane 70 |
| chr12_+_2170630 | 34.18 |
ENSRNOT00000071928
|
Pet100
|
PET100 homolog |
| chr2_+_218951141 | 34.16 |
ENSRNOT00000091001
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr2_+_84678948 | 34.14 |
ENSRNOT00000046325
|
Fam173b
|
family with sequence similarity 173, member B |
| chr8_+_117246376 | 34.10 |
ENSRNOT00000074493
|
Ccdc71
|
coiled-coil domain containing 71 |
| chr5_-_57267002 | 33.85 |
ENSRNOT00000011455
|
Bag1
|
Bcl2 associated athanogene 1 |
| chr6_+_2969333 | 33.79 |
ENSRNOT00000047356
|
Morn2
|
MORN repeat containing 2 |
| chr13_-_88536728 | 33.76 |
ENSRNOT00000003950
|
Uhmk1
|
U2AF homology motif kinase 1 |
| chr10_-_40201992 | 33.61 |
ENSRNOT00000075311
|
Lyrm7
|
LYR motif containing 7 |
| chr1_+_226947105 | 33.61 |
ENSRNOT00000028373
|
Prpf19
|
pre-mRNA processing factor 19 |
| chr3_+_61756148 | 33.49 |
ENSRNOT00000002134
|
Mtx2
|
metaxin 2 |
| chr1_-_222167447 | 33.41 |
ENSRNOT00000028687
|
Prdx5
|
peroxiredoxin 5 |
| chr5_-_134927235 | 33.23 |
ENSRNOT00000016751
|
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr7_+_121480723 | 33.16 |
ENSRNOT00000065304
|
Atf4
|
activating transcription factor 4 |
| chr1_-_72311856 | 33.10 |
ENSRNOT00000021286
|
Epn1
|
Epsin 1 |
| chr13_-_109483868 | 32.88 |
ENSRNOT00000067017
ENSRNOT00000087305 |
Rps6kc1
|
ribosomal protein S6 kinase C1 |
| chr3_-_161272460 | 32.79 |
ENSRNOT00000020740
|
Acot8
|
acyl-CoA thioesterase 8 |
| chr18_-_60002529 | 32.43 |
ENSRNOT00000081014
ENSRNOT00000024264 ENSRNOT00000059162 |
Nars
|
asparaginyl-tRNA synthetase |
| chr10_-_37209881 | 32.42 |
ENSRNOT00000090475
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chrX_-_32355296 | 32.21 |
ENSRNOT00000081652
ENSRNOT00000065075 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr9_+_17225122 | 32.19 |
ENSRNOT00000068747
ENSRNOT00000080816 |
Rsph9
|
radial spoke head 9 homolog |
| chr8_+_62696720 | 32.07 |
ENSRNOT00000010083
|
Ubl7
|
ubiquitin-like 7 |
| chr9_+_102862890 | 32.04 |
ENSRNOT00000050494
ENSRNOT00000080129 |
Fam174a
|
family with sequence similarity 174, member A |
| chr14_-_104375649 | 31.61 |
ENSRNOT00000006607
|
Actr2
|
ARP2 actin related protein 2 homolog |
| chr5_+_156668712 | 31.18 |
ENSRNOT00000067228
|
Ddost
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase non-catalytic subunit |
| chr5_+_172488708 | 31.15 |
ENSRNOT00000019930
|
Morn1
|
MORN repeat containing 1 |
| chr11_-_31103520 | 31.15 |
ENSRNOT00000030279
|
RGD1306954
|
similar to RIKEN cDNA 1110004E09 |
| chr3_-_22952063 | 31.02 |
ENSRNOT00000083850
|
Psmb7
|
proteasome subunit beta 7 |
| chr1_-_84986581 | 31.01 |
ENSRNOT00000025819
|
Psmc4
|
proteasome 26S subunit, ATPase 4 |
| chr6_+_86651196 | 30.83 |
ENSRNOT00000034756
|
RGD1307621
|
hypothetical LOC314168 |
| chr8_+_55037750 | 30.83 |
ENSRNOT00000013188
|
Timm8b
|
translocase of inner mitochondrial membrane 8 homolog B |
| chr1_+_170471238 | 30.73 |
ENSRNOT00000076961
ENSRNOT00000075597 ENSRNOT00000076631 ENSRNOT00000076783 |
Timm10b
Dnhd1
|
translocase of inner mitochondrial membrane 10B dynein heavy chain domain 1 |
| chr1_-_219259448 | 30.49 |
ENSRNOT00000024517
|
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr1_+_81365138 | 30.44 |
ENSRNOT00000026793
|
Cadm4
|
cell adhesion molecule 4 |
| chr3_+_113423693 | 30.18 |
ENSRNOT00000021091
|
Hypk
|
Huntingtin interacting protein K |
| chr1_-_170471076 | 30.18 |
ENSRNOT00000025159
|
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chr1_+_276309927 | 30.13 |
ENSRNOT00000067460
ENSRNOT00000066236 |
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr4_+_145399913 | 30.10 |
ENSRNOT00000012177
|
Jagn1
|
jagunal homolog 1 |
| chr5_+_162183327 | 30.10 |
ENSRNOT00000037875
|
Pramef8
|
PRAME family member 8 |
| chr8_+_112594691 | 29.93 |
ENSRNOT00000038383
ENSRNOT00000081281 |
Acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr6_+_108076306 | 29.66 |
ENSRNOT00000014913
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr1_-_192025350 | 29.66 |
ENSRNOT00000071946
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr1_-_92069160 | 29.57 |
ENSRNOT00000039115
|
Dpy19l3
|
dpy-19-like 3 (C. elegans) |
| chr3_-_1868015 | 29.56 |
ENSRNOT00000048945
|
Cacna1b
|
calcium voltage-gated channel subunit alpha1 B |
| chr9_-_44024876 | 29.41 |
ENSRNOT00000024366
|
Coa5
|
cytochrome C oxidase assembly factor 5 |
| chr2_-_20919824 | 29.33 |
ENSRNOT00000086300
|
Zcchc9
|
zinc finger CCHC-type containing 9 |
| chr16_-_2367069 | 29.29 |
ENSRNOT00000079625
|
Pde12
|
phosphodiesterase 12 |
| chr8_-_70215719 | 29.22 |
ENSRNOT00000015598
|
Rab11a
|
RAB11a, member RAS oncogene family |
| chr1_+_174655939 | 29.04 |
ENSRNOT00000014002
|
Ipo7
|
importin 7 |
| chr11_-_86328469 | 29.03 |
ENSRNOT00000071493
|
Ufd1l
|
ubiquitin fusion degradation 1 like (yeast) |
| chr8_+_117455262 | 29.02 |
ENSRNOT00000027520
|
Slc25a20
|
solute carrier family 25 member 20 |
| chrX_+_1787266 | 28.95 |
ENSRNOT00000011183
|
Ndufb11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr2_-_219842986 | 28.94 |
ENSRNOT00000055735
|
Agl
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr4_-_157266018 | 28.91 |
ENSRNOT00000019570
|
LOC100911713
|
protein C10-like |
| chr14_-_42520020 | 28.90 |
ENSRNOT00000065550
|
Slc30a9
|
solute carrier family 30 member 9 |
| chr8_-_114853103 | 28.82 |
ENSRNOT00000074595
|
Glyctk
|
glycerate kinase |
| chr11_-_24294179 | 28.79 |
ENSRNOT00000002116
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr3_+_121660110 | 28.70 |
ENSRNOT00000024942
|
Chchd5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr8_-_36410612 | 28.60 |
ENSRNOT00000091308
|
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr1_-_264756546 | 28.48 |
ENSRNOT00000020020
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr10_+_95706685 | 28.46 |
ENSRNOT00000004283
|
Psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr8_+_115179893 | 28.37 |
ENSRNOT00000017469
|
Rrp9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr8_+_61080103 | 28.29 |
ENSRNOT00000022976
|
Hmg20a
|
high mobility group 20A |
| chr8_-_77992621 | 28.22 |
ENSRNOT00000085843
|
Myzap
|
myocardial zonula adherens protein |
| chr6_-_26241337 | 28.15 |
ENSRNOT00000006525
|
Slc4a1ap
|
solute carrier family 4 member 1 adaptor protein |
| chr13_-_45127815 | 28.14 |
ENSRNOT00000005127
|
Dars
|
aspartyl-tRNA synthetase |
| chr5_-_136541795 | 28.08 |
ENSRNOT00000026336
|
Dmap1
|
DNA methyltransferase 1-associated protein 1 |
| chr2_-_188471988 | 28.08 |
ENSRNOT00000027785
|
Hcn3
|
hyperpolarization-activated cyclic nucleotide-gated potassium channel 3 |
| chr1_-_226572349 | 28.01 |
ENSRNOT00000028028
|
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr7_-_70355619 | 27.89 |
ENSRNOT00000031272
|
Tspan31
|
tetraspanin 31 |
| chr16_-_20873344 | 27.79 |
ENSRNOT00000027381
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr8_-_22625959 | 27.74 |
ENSRNOT00000012138
|
Yipf2
|
Yip1 domain family, member 2 |
| chr1_+_255479261 | 27.71 |
ENSRNOT00000079808
|
Tnks2
|
tankyrase 2 |
| chr9_+_43259709 | 27.68 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr1_+_84361736 | 27.62 |
ENSRNOT00000077968
|
RGD1307554
|
similar to CG16812-PA |
| chr12_-_48663143 | 27.55 |
ENSRNOT00000082809
|
Ficd
|
FIC domain containing |
| chr1_+_228142778 | 27.50 |
ENSRNOT00000028517
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr2_+_98252925 | 27.49 |
ENSRNOT00000011579
|
Pex2
|
peroxisomal biogenesis factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Etv3_Erf_Fev_Elk4_Elk1_Elk3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 42.3 | 169.3 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 37.9 | 113.7 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 25.1 | 75.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 22.0 | 131.8 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 21.2 | 106.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 19.5 | 58.5 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 16.6 | 33.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 16.5 | 82.6 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 15.2 | 76.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 15.2 | 45.5 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 15.2 | 45.5 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 14.9 | 164.4 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 14.5 | 43.5 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 13.9 | 41.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 13.4 | 40.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 13.2 | 39.7 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 13.0 | 77.8 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 12.8 | 38.3 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) fatty-acyl-CoA catabolic process(GO:0036115) |
| 12.1 | 48.6 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 12.1 | 72.7 | GO:1903215 | negative regulation of mRNA modification(GO:0090367) negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 11.8 | 58.9 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 11.2 | 44.9 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 11.1 | 33.4 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 10.8 | 32.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 10.4 | 83.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 9.4 | 18.7 | GO:0014736 | negative regulation of muscle atrophy(GO:0014736) |
| 9.1 | 27.4 | GO:1905244 | negative regulation of long term synaptic depression(GO:1900453) regulation of intracellular transport of viral material(GO:1901252) regulation of modification of synaptic structure(GO:1905244) |
| 9.0 | 116.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 8.5 | 34.0 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 8.3 | 25.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 8.0 | 24.0 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 7.9 | 79.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 7.9 | 23.6 | GO:0021678 | third ventricle development(GO:0021678) |
| 7.8 | 141.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 7.4 | 96.6 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 7.4 | 29.7 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 7.4 | 66.5 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 7.4 | 29.6 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 7.4 | 58.9 | GO:0051182 | coenzyme transport(GO:0051182) |
| 7.3 | 29.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 7.1 | 42.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 7.0 | 35.2 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 6.9 | 34.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 6.9 | 27.6 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 6.8 | 33.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 6.7 | 40.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 6.4 | 19.2 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 6.4 | 19.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 6.4 | 114.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 6.3 | 18.8 | GO:0060816 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 6.2 | 24.9 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 6.1 | 18.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 6.1 | 66.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 6.0 | 42.3 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 6.0 | 30.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 6.0 | 41.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 5.8 | 81.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 5.7 | 17.2 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 5.5 | 22.2 | GO:0051031 | tRNA transport(GO:0051031) |
| 5.5 | 21.9 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 5.5 | 16.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 5.4 | 16.3 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 5.4 | 123.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 5.3 | 16.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 5.2 | 15.7 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 5.1 | 40.8 | GO:0016559 | peroxisome fission(GO:0016559) |
| 5.1 | 5.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 4.9 | 14.6 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 4.9 | 19.4 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 4.8 | 43.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 4.8 | 28.6 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 4.7 | 47.4 | GO:0001302 | replicative cell aging(GO:0001302) |
| 4.7 | 9.5 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 4.7 | 9.4 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 4.7 | 174.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 4.7 | 18.7 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 4.6 | 13.9 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 4.6 | 55.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 4.6 | 23.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 4.6 | 18.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 4.5 | 13.6 | GO:0019249 | lactate biosynthetic process(GO:0019249) amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 4.5 | 76.9 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 4.3 | 39.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 4.3 | 51.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 4.2 | 12.7 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 4.2 | 42.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 4.2 | 12.6 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 4.2 | 12.6 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 4.1 | 20.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 4.1 | 12.4 | GO:0019677 | NADP catabolic process(GO:0006742) NAD catabolic process(GO:0019677) |
| 4.1 | 41.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 4.1 | 20.5 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 4.1 | 32.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 4.0 | 4.0 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 4.0 | 11.9 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 3.9 | 7.9 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 3.9 | 50.9 | GO:0090169 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 3.9 | 15.6 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 3.8 | 19.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 3.8 | 23.0 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 3.8 | 45.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 3.7 | 33.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 3.7 | 121.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 3.6 | 10.8 | GO:0043928 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 3.6 | 3.6 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 3.5 | 6.9 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 3.5 | 20.7 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 3.4 | 131.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 3.4 | 6.8 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 3.4 | 17.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 3.4 | 10.2 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 3.4 | 60.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 3.3 | 89.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 3.2 | 38.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 3.2 | 9.5 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 3.1 | 9.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 3.1 | 6.3 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 3.1 | 12.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 3.1 | 9.3 | GO:1903445 | protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 3.1 | 12.3 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 3.0 | 23.9 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 3.0 | 91.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 2.9 | 8.8 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 2.9 | 14.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 2.9 | 57.7 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 2.8 | 14.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 2.8 | 59.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 2.8 | 45.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 2.8 | 5.6 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 2.8 | 47.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 2.8 | 5.5 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 2.7 | 27.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 2.7 | 8.2 | GO:2000437 | regulation of monocyte extravasation(GO:2000437) |
| 2.7 | 29.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 2.7 | 32.6 | GO:0072718 | response to cisplatin(GO:0072718) |
| 2.7 | 5.4 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 2.7 | 8.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 2.7 | 61.3 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 2.7 | 66.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 2.6 | 18.3 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 2.6 | 26.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 2.6 | 38.9 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 2.6 | 10.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 2.6 | 25.8 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 2.6 | 7.7 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 2.6 | 30.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 2.6 | 10.3 | GO:0032439 | endosome localization(GO:0032439) |
| 2.5 | 10.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 2.5 | 22.5 | GO:0015825 | L-serine transport(GO:0015825) |
| 2.5 | 22.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 2.4 | 198.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 2.4 | 92.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 2.4 | 9.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 2.3 | 34.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 2.3 | 16.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 2.3 | 18.3 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) |
| 2.3 | 25.1 | GO:0042407 | cristae formation(GO:0042407) |
| 2.3 | 6.8 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 2.3 | 36.3 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 2.2 | 6.7 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 2.2 | 8.9 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 2.2 | 6.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 2.2 | 6.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 2.2 | 32.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 2.1 | 30.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 2.1 | 8.5 | GO:0061534 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 2.1 | 36.0 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 2.1 | 41.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 2.1 | 2.1 | GO:1904638 | response to resveratrol(GO:1904638) |
| 2.1 | 14.5 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 2.1 | 14.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 2.0 | 14.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 2.0 | 12.1 | GO:0097491 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 2.0 | 8.1 | GO:1903333 | negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 2.0 | 12.0 | GO:0042713 | sperm ejaculation(GO:0042713) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 2.0 | 23.8 | GO:0071806 | protein transmembrane transport(GO:0071806) |
| 2.0 | 7.9 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 2.0 | 5.9 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 2.0 | 5.9 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 2.0 | 23.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 1.9 | 30.9 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 1.9 | 15.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.9 | 9.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 1.9 | 85.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 1.9 | 28.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 1.9 | 13.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 1.9 | 14.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.8 | 9.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 1.8 | 32.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 1.8 | 40.0 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 1.8 | 3.6 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 1.8 | 17.7 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 1.7 | 5.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 1.7 | 85.6 | GO:0021762 | substantia nigra development(GO:0021762) |
| 1.7 | 5.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 1.6 | 9.9 | GO:0015074 | DNA integration(GO:0015074) |
| 1.6 | 9.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 1.6 | 8.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 1.6 | 16.1 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 1.6 | 4.8 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 1.6 | 9.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 1.6 | 12.6 | GO:0048278 | vesicle docking(GO:0048278) |
| 1.6 | 37.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 1.5 | 7.7 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 1.5 | 51.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 1.5 | 5.9 | GO:1990839 | response to endothelin(GO:1990839) cellular response to endothelin(GO:1990859) |
| 1.5 | 16.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 1.4 | 15.7 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 1.4 | 1.4 | GO:0046070 | dGTP metabolic process(GO:0046070) |
| 1.4 | 24.1 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 1.4 | 4.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 1.4 | 2.8 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 1.4 | 29.9 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 1.4 | 20.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 1.3 | 9.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 1.3 | 14.7 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 1.3 | 5.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 1.3 | 3.9 | GO:0007228 | signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 1.3 | 2.6 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 1.3 | 7.6 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 1.2 | 8.7 | GO:0060526 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 1.2 | 5.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 1.2 | 38.9 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 1.2 | 4.8 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 1.2 | 2.4 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 1.2 | 10.8 | GO:0007000 | nucleolus organization(GO:0007000) |
| 1.2 | 158.7 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 1.2 | 16.5 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.2 | 36.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 1.2 | 3.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 1.2 | 2.3 | GO:0032258 | CVT pathway(GO:0032258) |
| 1.1 | 33.8 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 1.1 | 3.4 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 1.1 | 6.7 | GO:1901621 | response to clozapine(GO:0097338) negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 1.1 | 10.8 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 1.1 | 7.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 1.1 | 4.2 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 1.1 | 54.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 1.1 | 10.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 1.0 | 9.3 | GO:2000650 | negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 1.0 | 3.1 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 1.0 | 10.2 | GO:0006183 | GTP biosynthetic process(GO:0006183) guanosine-containing compound biosynthetic process(GO:1901070) |
| 1.0 | 13.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 1.0 | 18.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 1.0 | 12.4 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.9 | 20.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.9 | 42.8 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.9 | 46.8 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.9 | 23.8 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.9 | 6.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.9 | 2.6 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.9 | 8.6 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.8 | 32.2 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.8 | 3.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.8 | 4.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.8 | 8.2 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.8 | 4.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.8 | 25.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) tRNA aminoacylation(GO:0043039) |
| 0.8 | 16.7 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.8 | 1.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.8 | 7.7 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.8 | 7.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.8 | 6.9 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.8 | 15.2 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.8 | 1.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.7 | 4.5 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.7 | 2.2 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.7 | 5.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.7 | 5.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.7 | 2.2 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) mammary gland specification(GO:0060594) |
| 0.7 | 5.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.7 | 1.4 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.7 | 9.9 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.7 | 35.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.7 | 4.9 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.7 | 9.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.7 | 8.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.7 | 13.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.7 | 12.9 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.7 | 7.5 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.7 | 20.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.7 | 26.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) synaptic transmission, cholinergic(GO:0007271) |
| 0.7 | 5.3 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.7 | 11.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.7 | 41.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.6 | 6.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.6 | 11.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.6 | 3.8 | GO:0021764 | amygdala development(GO:0021764) |
| 0.6 | 28.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.6 | 3.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.6 | 0.6 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.6 | 20.8 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.6 | 12.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.6 | 7.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.6 | 2.4 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.6 | 0.6 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.6 | 5.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.6 | 4.6 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.6 | 13.9 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.6 | 4.0 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.6 | 1.7 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.6 | 26.2 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.5 | 6.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.5 | 7.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.5 | 8.5 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.5 | 15.8 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.5 | 5.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.5 | 20.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.5 | 8.0 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.5 | 2.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.5 | 3.5 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.5 | 6.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.5 | 11.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.5 | 2.4 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.5 | 1.9 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.5 | 1.4 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.4 | 112.0 | GO:0010498 | proteasomal protein catabolic process(GO:0010498) |
| 0.4 | 1.8 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.4 | 5.8 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.4 | 33.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.4 | 27.4 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.4 | 9.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 3.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.4 | 20.7 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.4 | 2.1 | GO:0090494 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.4 | 19.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.4 | 0.8 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.4 | 3.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 10.9 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) |
| 0.4 | 6.8 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.4 | 7.5 | GO:0097480 | synaptic vesicle transport(GO:0048489) establishment of synaptic vesicle localization(GO:0097480) |
| 0.4 | 3.0 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.4 | 0.7 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.4 | 12.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.4 | 2.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.4 | 14.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.4 | 4.9 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.4 | 4.9 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.3 | 2.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 1.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 1.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 2.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.3 | 7.6 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.3 | 3.4 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.3 | 2.1 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 1.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 0.9 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.3 | 2.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.3 | 5.8 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.3 | 0.6 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.3 | 6.2 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.3 | 2.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.3 | 0.8 | GO:0015746 | citrate transport(GO:0015746) |
| 0.3 | 13.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 0.5 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.2 | 16.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.2 | 2.4 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.2 | 3.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 0.7 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.2 | 3.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 8.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.2 | 56.1 | GO:0006887 | exocytosis(GO:0006887) |
| 0.2 | 21.6 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.2 | 0.6 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.2 | 1.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.2 | 8.8 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.2 | 2.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.2 | 2.0 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.2 | 25.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 6.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.2 | 2.1 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 6.4 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.2 | 5.7 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.2 | 19.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.2 | 2.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 1.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.2 | 0.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 4.9 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.2 | 3.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 8.9 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.1 | 2.3 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.1 | 11.1 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 2.6 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 0.7 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 3.8 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 1.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 2.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 2.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 1.4 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 2.4 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.1 | 2.8 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.7 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 6.3 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.1 | 2.3 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 0.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 8.8 | GO:0050890 | cognition(GO:0050890) |
| 0.1 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 3.2 | GO:0003014 | renal system process(GO:0003014) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 1.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 38.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.9 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 47.5 | 142.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 44.1 | 176.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 27.7 | 83.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 19.5 | 78.0 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 17.9 | 143.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 14.8 | 59.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 14.2 | 42.5 | GO:0005962 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 13.9 | 41.8 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 13.6 | 109.0 | GO:0016272 | prefoldin complex(GO:0016272) |
| 13.3 | 40.0 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 11.7 | 58.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 11.2 | 55.9 | GO:0097361 | CIA complex(GO:0097361) |
| 11.1 | 33.2 | GO:1990589 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 10.8 | 32.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 10.5 | 146.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 10.3 | 51.7 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 9.1 | 27.4 | GO:0097444 | spine apparatus(GO:0097444) |
| 9.1 | 36.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 8.7 | 34.7 | GO:0045281 | respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) |
| 8.4 | 25.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 8.3 | 33.1 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 7.6 | 45.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 7.2 | 36.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 7.2 | 368.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 7.2 | 100.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 7.2 | 42.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 6.9 | 20.6 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 6.8 | 88.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 6.6 | 39.7 | GO:0089701 | U2AF(GO:0089701) |
| 6.6 | 46.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 6.6 | 131.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 6.6 | 46.1 | GO:0070449 | elongin complex(GO:0070449) |
| 6.2 | 43.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 6.1 | 61.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 5.7 | 68.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 5.7 | 34.1 | GO:0030478 | actin cap(GO:0030478) |
| 5.6 | 28.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 5.6 | 44.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 5.6 | 83.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 5.5 | 32.8 | GO:0070847 | core mediator complex(GO:0070847) |
| 5.4 | 10.8 | GO:0055087 | Ski complex(GO:0055087) |
| 5.3 | 78.9 | GO:0034709 | methylosome(GO:0034709) |
| 5.2 | 151.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 5.2 | 46.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 5.1 | 15.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 5.0 | 15.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 4.6 | 63.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 4.4 | 13.3 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 4.4 | 57.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 4.4 | 69.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 4.2 | 12.6 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 4.2 | 92.0 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 4.0 | 12.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 4.0 | 27.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 3.9 | 232.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 3.9 | 46.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 3.8 | 30.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 3.8 | 41.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 3.8 | 30.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 3.7 | 29.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 3.6 | 10.9 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 3.6 | 32.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 3.6 | 81.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 3.4 | 51.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 3.4 | 80.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 3.4 | 6.7 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 3.3 | 42.8 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 3.2 | 29.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 3.2 | 19.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 3.2 | 32.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 3.2 | 16.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 3.2 | 25.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 3.1 | 12.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 3.1 | 12.4 | GO:0034657 | GID complex(GO:0034657) |
| 3.1 | 21.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 3.1 | 18.3 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 3.0 | 33.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 2.9 | 11.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 2.9 | 5.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 2.7 | 101.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 2.6 | 18.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 2.6 | 33.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 2.5 | 20.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 2.5 | 17.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 2.4 | 4.8 | GO:0097413 | Lewy body(GO:0097413) |
| 2.3 | 41.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 2.3 | 13.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 2.1 | 8.6 | GO:0032021 | NELF complex(GO:0032021) |
| 2.1 | 12.8 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 2.1 | 26.9 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 2.1 | 45.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 2.0 | 28.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 2.0 | 16.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 2.0 | 10.0 | GO:0070069 | cytochrome complex(GO:0070069) |
| 2.0 | 31.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 1.9 | 5.8 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 1.9 | 9.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 1.9 | 37.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 1.8 | 9.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 1.8 | 9.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 1.8 | 43.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 1.8 | 8.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 1.8 | 8.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.8 | 28.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 1.6 | 16.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 1.6 | 8.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 1.6 | 6.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 1.6 | 43.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 1.6 | 82.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 1.6 | 11.0 | GO:0048500 | signal recognition particle(GO:0048500) |
| 1.5 | 16.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 1.5 | 92.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 1.5 | 27.9 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 1.5 | 10.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.5 | 59.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 1.4 | 28.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 1.4 | 30.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 1.3 | 14.8 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 1.3 | 11.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 1.3 | 31.7 | GO:0000145 | exocyst(GO:0000145) |
| 1.3 | 736.1 | GO:0044429 | mitochondrial part(GO:0044429) |
| 1.2 | 15.0 | GO:0032797 | SMN complex(GO:0032797) |
| 1.2 | 5.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.2 | 13.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 1.2 | 13.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 1.2 | 57.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 1.1 | 11.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 1.1 | 2.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 1.1 | 26.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 1.0 | 45.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 1.0 | 22.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.0 | 16.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.9 | 5.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.9 | 14.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.9 | 6.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.9 | 5.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.9 | 7.0 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.9 | 19.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.9 | 39.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.9 | 2.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.9 | 5.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.8 | 5.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.8 | 12.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.8 | 58.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.8 | 4.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.8 | 2.4 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.8 | 8.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.8 | 8.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.7 | 7.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.7 | 6.7 | GO:0032059 | bleb(GO:0032059) |
| 0.7 | 6.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.6 | 27.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.6 | 33.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.6 | 54.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.6 | 19.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.6 | 5.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.6 | 4.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.6 | 8.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.6 | 42.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.6 | 10.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.6 | 3.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.5 | 8.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.5 | 12.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.5 | 21.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.5 | 8.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.5 | 3.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.5 | 11.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.4 | 7.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.4 | 31.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.4 | 20.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.4 | 19.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.4 | 4.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.4 | 7.9 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.4 | 1.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.4 | 71.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.4 | 18.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 3.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 3.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 3.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.3 | 308.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.3 | 16.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 5.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.3 | 1.5 | GO:0035859 | Seh1-associated complex(GO:0035859) |
| 0.3 | 1.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.3 | 2.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 1.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 1.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.3 | 66.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.3 | 3.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.3 | 20.1 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.3 | 9.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 228.4 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.2 | 22.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 13.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 19.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 0.7 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.2 | 14.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 3.4 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.2 | 11.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 27.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.2 | 3.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 1.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 3.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 3.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 0.9 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 13.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 17.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 1.6 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.2 | 3.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 28.3 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.1 | 4.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 15.2 | GO:0044431 | Golgi apparatus part(GO:0044431) |
| 0.1 | 397.9 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.5 | GO:1902554 | serine/threonine protein kinase complex(GO:1902554) |
| 0.0 | 1.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.0 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 25.1 | 75.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 24.1 | 96.5 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 23.8 | 71.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 19.5 | 58.5 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 19.1 | 114.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 18.9 | 75.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 18.6 | 55.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 16.4 | 81.8 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 16.2 | 64.7 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 14.4 | 43.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 14.4 | 231.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 13.9 | 41.8 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 13.5 | 148.9 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 13.3 | 40.0 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 12.7 | 38.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 12.3 | 37.0 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 10.6 | 42.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 10.5 | 52.6 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 10.0 | 29.9 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 9.7 | 29.0 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 9.5 | 47.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 9.3 | 74.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 9.2 | 248.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 8.8 | 35.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 8.7 | 34.7 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 8.4 | 33.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 7.9 | 62.9 | GO:0071253 | connexin binding(GO:0071253) |
| 6.8 | 27.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 6.7 | 26.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 6.6 | 26.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 6.4 | 19.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 6.4 | 19.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 6.4 | 44.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 6.3 | 25.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 6.2 | 37.0 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 6.1 | 49.1 | GO:0031386 | protein tag(GO:0031386) |
| 5.7 | 17.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 5.6 | 78.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 5.6 | 16.7 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 5.5 | 49.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 5.4 | 54.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 5.4 | 27.1 | GO:0089720 | caspase binding(GO:0089720) |
| 5.2 | 20.8 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 5.2 | 62.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 5.2 | 62.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 5.1 | 45.5 | GO:0043495 | protein anchor(GO:0043495) |
| 5.0 | 40.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 4.9 | 108.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 4.8 | 33.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 4.7 | 56.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 4.7 | 18.7 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 4.5 | 13.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 4.5 | 17.8 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 4.4 | 30.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 4.3 | 34.7 | GO:0034046 | poly(G) binding(GO:0034046) |
| 4.3 | 17.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 4.2 | 17.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 4.2 | 20.8 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 4.2 | 33.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 4.1 | 12.3 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 4.1 | 53.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 4.0 | 39.7 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 3.9 | 11.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 3.8 | 38.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 3.7 | 14.8 | GO:0034618 | arginine binding(GO:0034618) |
| 3.7 | 99.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 3.7 | 14.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 3.6 | 39.3 | GO:0070513 | death domain binding(GO:0070513) |
| 3.6 | 107.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 3.5 | 10.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 3.4 | 10.3 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 3.4 | 304.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 3.4 | 13.7 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 3.3 | 13.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 3.3 | 72.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 3.2 | 16.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 3.2 | 25.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 3.2 | 66.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 3.1 | 31.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 3.1 | 12.4 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 3.1 | 42.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 3.0 | 12.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 2.9 | 20.6 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 2.8 | 8.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.8 | 34.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 2.7 | 19.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 2.7 | 10.7 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 2.6 | 34.0 | GO:0008061 | chitin binding(GO:0008061) |
| 2.6 | 15.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 2.5 | 12.7 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 2.5 | 7.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 2.5 | 74.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 2.5 | 37.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 2.5 | 9.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 2.4 | 19.4 | GO:0046790 | virion binding(GO:0046790) |
| 2.4 | 2.4 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 2.3 | 13.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 2.3 | 31.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 2.2 | 6.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 2.1 | 108.1 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 2.1 | 8.5 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 2.0 | 32.3 | GO:0046933 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 2.0 | 14.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 2.0 | 12.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 2.0 | 6.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 2.0 | 27.9 | GO:0000339 | RNA cap binding(GO:0000339) |
| 2.0 | 19.9 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 1.9 | 15.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.9 | 188.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 1.9 | 5.6 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 1.8 | 11.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.8 | 29.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 1.8 | 5.4 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 1.8 | 12.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 1.7 | 29.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 1.7 | 5.1 | GO:0001031 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) 5S rDNA binding(GO:0080084) |
| 1.7 | 11.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 1.6 | 16.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 1.6 | 75.4 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 1.6 | 29.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.5 | 26.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 1.5 | 63.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 1.5 | 5.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.4 | 10.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 1.3 | 20.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 1.3 | 7.9 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.3 | 58.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 1.3 | 10.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.3 | 3.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 1.3 | 38.9 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 1.2 | 13.7 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 1.2 | 8.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 1.2 | 4.8 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 1.2 | 79.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 1.2 | 24.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 1.2 | 9.6 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 1.2 | 26.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 1.2 | 8.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.1 | 29.7 | GO:0071949 | FAD binding(GO:0071949) |
| 1.1 | 4.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 1.1 | 9.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 1.1 | 4.4 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 1.1 | 8.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.1 | 3.2 | GO:0019961 | interferon binding(GO:0019961) |
| 1.0 | 9.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 1.0 | 2.1 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 1.0 | 8.3 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 1.0 | 18.3 | GO:0005522 | profilin binding(GO:0005522) |
| 1.0 | 8.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 1.0 | 5.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.0 | 4.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 1.0 | 5.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 1.0 | 6.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 1.0 | 14.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.9 | 5.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.9 | 110.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.9 | 30.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.9 | 31.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.9 | 25.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.9 | 7.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.9 | 38.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.9 | 12.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.9 | 17.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.9 | 2.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.9 | 6.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.9 | 4.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.9 | 20.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.8 | 31.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.8 | 0.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.8 | 5.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.8 | 4.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.8 | 14.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.8 | 116.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.8 | 7.6 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.8 | 3.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.7 | 9.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.7 | 3.7 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.7 | 20.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.7 | 8.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.7 | 2.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.7 | 26.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.7 | 2.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.7 | 6.9 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.7 | 13.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.7 | 0.7 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.7 | 2.0 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.7 | 7.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.7 | 2.0 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 0.7 | 27.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.6 | 20.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.6 | 6.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.6 | 180.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.6 | 0.6 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.6 | 8.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.6 | 1.7 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.6 | 4.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.6 | 2.2 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.6 | 2.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.5 | 189.5 | GO:0019787 | ubiquitin-like protein transferase activity(GO:0019787) |
| 0.5 | 20.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.5 | 15.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.5 | 7.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.5 | 115.4 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.5 | 3.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 3.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 6.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.5 | 26.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.5 | 4.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.5 | 2.8 | GO:0016531 | copper chaperone activity(GO:0016531) cuprous ion binding(GO:1903136) |
| 0.4 | 3.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 6.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.4 | 23.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.4 | 5.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.4 | 5.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 7.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.4 | 9.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.4 | 9.9 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.4 | 10.8 | GO:0043531 | ADP binding(GO:0043531) |
| 0.4 | 0.8 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 0.4 | 14.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.4 | 3.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.4 | 7.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.4 | 70.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.4 | 6.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.4 | 6.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 3.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.4 | 8.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.4 | 1.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.3 | 5.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 50.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.3 | 1.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 2.3 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.3 | 246.8 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.3 | 3.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 5.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.3 | 7.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 66.8 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.3 | 2.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 1.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 1.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.3 | 6.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.3 | 16.6 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.3 | 0.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.3 | 66.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 101.6 | GO:0046873 | metal ion transmembrane transporter activity(GO:0046873) |
| 0.3 | 3.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 3.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 0.7 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 0.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 12.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.2 | 11.9 | GO:0015405 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) |
| 0.2 | 0.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 2.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 1.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 2.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 2.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 5.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 5.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 1.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 2.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 11.3 | GO:0042277 | peptide binding(GO:0042277) |
| 0.1 | 2.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 2.2 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.1 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 6.2 | GO:0020037 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.1 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 11.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.6 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.4 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 1.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.6 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 35.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 100.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 3.0 | 78.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 2.9 | 64.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 2.6 | 107.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 2.4 | 33.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 2.4 | 31.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 1.7 | 59.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 1.5 | 27.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 1.5 | 14.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.3 | 56.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 1.2 | 34.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 1.2 | 43.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 1.2 | 22.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 1.1 | 36.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 1.1 | 12.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 1.1 | 71.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 1.0 | 32.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.8 | 21.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.8 | 6.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.8 | 13.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.7 | 9.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.7 | 18.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.7 | 11.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.6 | 19.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.6 | 13.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.5 | 17.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.5 | 2.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.5 | 30.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.4 | 27.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.4 | 8.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.4 | 2.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.4 | 3.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.4 | 2.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.3 | 10.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 4.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 3.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 2.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 6.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 8.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 5.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 8.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 9.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 19.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 85.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 7.5 | 135.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 7.1 | 114.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 5.9 | 307.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 5.8 | 69.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 5.6 | 343.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 5.3 | 142.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 5.0 | 15.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 4.8 | 81.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 4.7 | 66.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 3.9 | 94.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 3.9 | 38.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 3.9 | 46.2 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 3.5 | 62.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 3.4 | 67.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 3.4 | 140.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 3.0 | 15.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 2.9 | 23.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 2.7 | 54.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 2.7 | 69.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 2.7 | 72.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 2.7 | 42.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 2.6 | 71.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 2.6 | 163.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 2.6 | 40.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 2.4 | 43.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 2.4 | 63.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 2.2 | 26.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 2.1 | 37.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 2.0 | 30.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 2.0 | 67.0 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 2.0 | 22.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 2.0 | 86.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 2.0 | 23.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 1.9 | 26.1 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 1.9 | 14.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 1.6 | 38.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 1.5 | 27.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 1.4 | 18.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 1.3 | 15.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 1.2 | 10.9 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 1.2 | 29.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 1.2 | 11.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 1.0 | 33.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 1.0 | 17.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.9 | 11.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.9 | 41.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.9 | 20.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.9 | 11.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.8 | 40.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.8 | 18.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.7 | 5.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.6 | 10.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.6 | 5.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.5 | 6.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.5 | 10.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.5 | 10.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.5 | 10.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.4 | 4.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.4 | 6.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.4 | 10.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.4 | 5.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.4 | 60.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.4 | 9.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 7.6 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.3 | 3.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 37.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.3 | 17.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.3 | 4.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.3 | 8.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 28.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.3 | 4.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 3.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 26.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 5.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 24.1 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.2 | 3.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 3.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 2.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 4.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.2 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.1 | 0.7 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 1.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 5.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 10.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |