Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
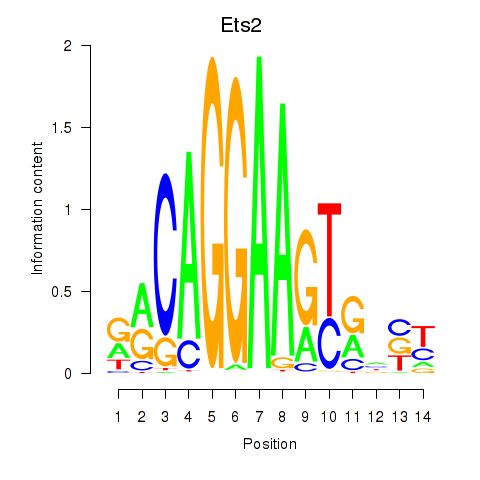
Results for Ets2
Z-value: 0.60
Transcription factors associated with Ets2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ets2
|
ENSRNOG00000001647 | ETS proto-oncogene 2, transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ets2 | rn6_v1_chr11_+_36075709_36075709 | 0.34 | 2.9e-10 | Click! |
Activity profile of Ets2 motif
Sorted Z-values of Ets2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_68360664 | 17.81 |
ENSRNOT00000030971
|
Hmcn1
|
hemicentin 1 |
| chr16_+_18716019 | 15.36 |
ENSRNOT00000047870
|
Sftpa1
|
surfactant protein A1 |
| chr6_-_79306443 | 13.39 |
ENSRNOT00000030706
|
Clec14a
|
C-type lectin domain family 14, member A |
| chr10_-_40764185 | 12.58 |
ENSRNOT00000017486
|
Sparc
|
secreted protein acidic and cysteine rich |
| chr1_+_81763614 | 11.78 |
ENSRNOT00000027254
|
Cd79a
|
CD79a molecule |
| chr7_+_142776252 | 11.35 |
ENSRNOT00000008673
|
Acvrl1
|
activin A receptor like type 1 |
| chr7_+_142776580 | 11.05 |
ENSRNOT00000081047
|
Acvrl1
|
activin A receptor like type 1 |
| chr2_-_190100276 | 10.97 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr7_-_107392972 | 10.95 |
ENSRNOT00000093425
|
Tmem71
|
transmembrane protein 71 |
| chr7_+_14037620 | 10.81 |
ENSRNOT00000009606
|
Syde1
|
synapse defective Rho GTPase homolog 1 |
| chr9_-_16795887 | 10.63 |
ENSRNOT00000071609
|
Cd79al
|
Cd79a molecule-like |
| chr4_+_78320190 | 10.61 |
ENSRNOT00000032742
ENSRNOT00000091359 |
Gimap4
|
GTPase, IMAP family member 4 |
| chr1_-_227441442 | 9.23 |
ENSRNOT00000028433
|
Ms4a1
|
membrane spanning 4-domains A1 |
| chr3_+_11679530 | 9.18 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr4_+_158088505 | 9.14 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr2_+_188745503 | 8.89 |
ENSRNOT00000056652
|
Shc1
|
SHC adaptor protein 1 |
| chr14_+_80195715 | 8.67 |
ENSRNOT00000010784
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr10_+_35133252 | 8.53 |
ENSRNOT00000051916
|
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr3_+_93909156 | 8.42 |
ENSRNOT00000090365
|
Lmo2
|
LIM domain only 2 |
| chr4_+_28989115 | 8.31 |
ENSRNOT00000075326
|
Gng11
|
G protein subunit gamma 11 |
| chr7_-_64251110 | 8.25 |
ENSRNOT00000006180
|
Srgap1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr2_-_187133993 | 8.24 |
ENSRNOT00000019502
|
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr1_-_221015929 | 8.21 |
ENSRNOT00000028137
|
Sipa1
|
signal-induced proliferation-associated 1 |
| chr1_-_175796040 | 8.15 |
ENSRNOT00000024060
|
Mrvi1
|
murine retrovirus integration site 1 homolog |
| chr4_-_78342863 | 7.96 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr4_+_31387420 | 7.79 |
ENSRNOT00000074048
|
LOC100912034
|
guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-11-like |
| chr14_+_34727915 | 7.77 |
ENSRNOT00000085089
|
Kdr
|
kinase insert domain receptor |
| chr5_-_59063592 | 7.72 |
ENSRNOT00000022400
|
Tln1
|
talin 1 |
| chr14_+_70780623 | 7.65 |
ENSRNOT00000083871
ENSRNOT00000058803 |
Ldb2
|
LIM domain binding 2 |
| chr7_-_118108864 | 7.55 |
ENSRNOT00000006184
|
Mb
|
myoglobin |
| chr9_-_27192135 | 7.54 |
ENSRNOT00000017592
|
Tram2
|
translocation associated membrane protein 2 |
| chr5_+_16526058 | 7.51 |
ENSRNOT00000011130
|
Lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr14_-_86796378 | 7.50 |
ENSRNOT00000092021
|
Myo1g
|
myosin IG |
| chr3_+_16610086 | 7.40 |
ENSRNOT00000046231
|
LOC100361009
|
rCG64257-like |
| chr10_-_70802782 | 7.35 |
ENSRNOT00000045867
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr15_+_57221292 | 7.34 |
ENSRNOT00000014502
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr1_-_199624783 | 7.02 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr4_-_170080456 | 7.01 |
ENSRNOT00000070936
|
LOC100912527
|
uncharacterized LOC100912527 |
| chr8_-_116465801 | 6.94 |
ENSRNOT00000085915
|
Sema3f
|
semaphorin 3F |
| chr2_-_62634785 | 6.91 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr16_+_10727571 | 6.87 |
ENSRNOT00000084422
|
Mmrn2
|
multimerin 2 |
| chr5_+_82587420 | 6.85 |
ENSRNOT00000014020
|
Tlr4
|
toll-like receptor 4 |
| chr10_+_94566928 | 6.82 |
ENSRNOT00000078446
|
Prr29
|
proline rich 29 |
| chr17_-_10001901 | 6.80 |
ENSRNOT00000082836
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr1_-_216918969 | 6.74 |
ENSRNOT00000079454
|
Osbpl5
|
oxysterol binding protein-like 5 |
| chr13_-_52514875 | 6.73 |
ENSRNOT00000064758
|
Nav1
|
neuron navigator 1 |
| chr9_-_119332967 | 6.73 |
ENSRNOT00000021048
|
Myl12a
|
myosin light chain 12A |
| chr1_+_88875375 | 6.64 |
ENSRNOT00000028284
|
Tyrobp
|
Tyro protein tyrosine kinase binding protein |
| chr1_-_89509343 | 6.43 |
ENSRNOT00000028637
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr8_+_39878955 | 6.43 |
ENSRNOT00000060300
|
LOC100911068
|
roundabout homolog 4-like |
| chr16_-_9430743 | 6.37 |
ENSRNOT00000043811
|
Wdfy4
|
WDFY family member 4 |
| chr5_-_145377221 | 6.23 |
ENSRNOT00000019246
|
Gja4
|
gap junction protein, alpha 4 |
| chr9_+_10952374 | 6.18 |
ENSRNOT00000074993
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr1_-_260254600 | 5.87 |
ENSRNOT00000019014
|
Blnk
|
B-cell linker |
| chr8_-_39093277 | 5.85 |
ENSRNOT00000043344
|
LOC100911068
|
roundabout homolog 4-like |
| chr6_+_98284170 | 5.84 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr10_+_63662986 | 5.77 |
ENSRNOT00000088722
|
Scarf1
|
scavenger receptor class F, member 1 |
| chr20_+_30915213 | 5.70 |
ENSRNOT00000000681
|
Prf1
|
perforin 1 |
| chr2_-_113616766 | 5.58 |
ENSRNOT00000016858
ENSRNOT00000074723 |
Tmem212
|
transmembrane protein 212 |
| chr3_+_122544788 | 5.54 |
ENSRNOT00000063828
|
Tgm3
|
transglutaminase 3 |
| chr4_-_170129407 | 5.52 |
ENSRNOT00000075717
|
LOC100912609
|
uncharacterized LOC100912609 |
| chr3_+_80555196 | 5.37 |
ENSRNOT00000067318
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr6_+_96479430 | 5.24 |
ENSRNOT00000006729
|
Prkch
|
protein kinase C, eta |
| chr10_+_56764927 | 5.08 |
ENSRNOT00000025308
|
Clec10a
|
C-type lectin domain family 10, member A |
| chr1_+_215610368 | 5.07 |
ENSRNOT00000078903
ENSRNOT00000087781 |
Tnni2
|
troponin I2, fast skeletal type |
| chrX_+_68752597 | 5.02 |
ENSRNOT00000077039
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr3_-_110517163 | 4.95 |
ENSRNOT00000078037
|
Plcb2
|
phospholipase C, beta 2 |
| chr10_+_59529785 | 4.93 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr15_-_34469350 | 4.89 |
ENSRNOT00000067536
|
Adcy4
|
adenylate cyclase 4 |
| chr8_+_116324040 | 4.87 |
ENSRNOT00000081353
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr8_+_107882219 | 4.80 |
ENSRNOT00000019902
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr3_+_58965552 | 4.72 |
ENSRNOT00000002068
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr1_+_81779380 | 4.72 |
ENSRNOT00000065865
ENSRNOT00000080143 ENSRNOT00000089592 ENSRNOT00000080840 |
Arhgef1
|
Rho guanine nucleotide exchange factor 1 |
| chr1_+_175586097 | 4.70 |
ENSRNOT00000024933
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr5_-_159119171 | 4.68 |
ENSRNOT00000074074
ENSRNOT00000075382 ENSRNOT00000074378 |
Arhgef10l
|
Rho guanine nucleotide exchange factor 10 like |
| chr1_-_242083484 | 4.57 |
ENSRNOT00000065921
|
Tjp2
|
tight junction protein 2 |
| chr18_-_56115593 | 4.44 |
ENSRNOT00000045041
|
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr9_+_42871950 | 4.41 |
ENSRNOT00000089673
|
Arid5a
|
AT-rich interaction domain 5A |
| chr1_-_89473904 | 4.41 |
ENSRNOT00000089474
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr7_+_12471824 | 4.33 |
ENSRNOT00000068197
|
Sbno2
|
strawberry notch homolog 2 |
| chr16_+_4469468 | 4.32 |
ENSRNOT00000021164
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr6_-_43448280 | 4.27 |
ENSRNOT00000081110
|
Adam17
|
ADAM metallopeptidase domain 17 |
| chr13_-_67206688 | 4.25 |
ENSRNOT00000003630
ENSRNOT00000090693 |
Pla2g4a
|
phospholipase A2 group IVA |
| chr20_+_46199981 | 4.22 |
ENSRNOT00000000337
|
Mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr13_-_88642011 | 4.21 |
ENSRNOT00000067037
|
LOC100361087
|
hypothetical LOC100361087 |
| chr14_+_1463359 | 4.20 |
ENSRNOT00000070834
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
| chr19_+_49016891 | 4.19 |
ENSRNOT00000016713
|
Dynlrb2
|
dynein light chain roadblock-type 2 |
| chr4_+_56625561 | 4.10 |
ENSRNOT00000008356
ENSRNOT00000090808 ENSRNOT00000008972 |
Calu
|
calumenin |
| chr1_-_219438779 | 4.06 |
ENSRNOT00000029237
|
Tbc1d10c
|
TBC1 domain family, member 10C |
| chr14_-_81023682 | 4.01 |
ENSRNOT00000081570
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr1_+_23409408 | 3.98 |
ENSRNOT00000022362
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr1_+_256786124 | 3.92 |
ENSRNOT00000034563
|
Ffar4
|
free fatty acid receptor 4 |
| chr10_-_13892997 | 3.88 |
ENSRNOT00000004192
|
Traf7
|
TNF receptor associated factor 7 |
| chr1_+_122981755 | 3.83 |
ENSRNOT00000013468
|
Ndn
|
necdin, MAGE family member |
| chr13_+_89774764 | 3.80 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr15_+_52451161 | 3.79 |
ENSRNOT00000018725
|
Dok2
|
docking protein 2 |
| chr1_-_89474252 | 3.75 |
ENSRNOT00000028597
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr5_-_160158386 | 3.73 |
ENSRNOT00000089345
|
Fblim1
|
filamin binding LIM protein 1 |
| chr20_+_13662867 | 3.71 |
ENSRNOT00000032257
|
RGD1564162
|
similar to Homo sapiens fetal lung specific expression unknown |
| chrX_-_32355296 | 3.65 |
ENSRNOT00000081652
ENSRNOT00000065075 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr11_-_14304603 | 3.62 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr2_+_187447501 | 3.57 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr1_-_169456098 | 3.57 |
ENSRNOT00000030827
|
Trim30c
|
tripartite motif-containing 30C |
| chr10_-_30118873 | 3.54 |
ENSRNOT00000006063
|
Ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr19_+_19395655 | 3.49 |
ENSRNOT00000019130
|
Snx20
|
sorting nexin 20 |
| chr8_-_72204730 | 3.46 |
ENSRNOT00000023810
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr5_-_137238354 | 3.45 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr5_+_58661049 | 3.44 |
ENSRNOT00000078274
|
Unc13b
|
unc-13 homolog B |
| chr20_+_6973398 | 3.35 |
ENSRNOT00000041665
|
Fgd2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr5_-_152458023 | 3.34 |
ENSRNOT00000031225
|
Cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr11_+_82680253 | 3.33 |
ENSRNOT00000077119
ENSRNOT00000075512 |
Liph
|
lipase H |
| chr10_-_62483342 | 3.32 |
ENSRNOT00000079956
|
LOC100912068
|
hypermethylated in cancer 1 protein-like |
| chr10_-_85084850 | 3.32 |
ENSRNOT00000012462
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr1_+_87938042 | 3.30 |
ENSRNOT00000027837
|
Map4k1
|
mitogen activated protein kinase kinase kinase kinase 1 |
| chrX_-_157474219 | 3.29 |
ENSRNOT00000079860
|
Zfp275
|
zinc finger protein 275 |
| chr16_+_23317953 | 3.22 |
ENSRNOT00000075287
|
AABR07024972.1
|
|
| chr13_+_52645257 | 3.22 |
ENSRNOT00000012801
|
Lad1
|
ladinin 1 |
| chr15_+_8730871 | 3.22 |
ENSRNOT00000081845
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr8_-_65587658 | 3.21 |
ENSRNOT00000091982
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr5_-_33182147 | 3.20 |
ENSRNOT00000080358
|
Maged2
|
MAGE family member D2 |
| chr2_-_189573280 | 3.19 |
ENSRNOT00000022897
|
Rps27
|
ribosomal protein S27 |
| chr7_-_73450262 | 3.15 |
ENSRNOT00000006943
|
Nipal2
|
NIPA-like domain containing 2 |
| chr10_+_74002151 | 3.10 |
ENSRNOT00000005661
|
Ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr9_-_113598477 | 3.05 |
ENSRNOT00000035606
ENSRNOT00000084884 |
Ralbp1
|
ralA binding protein 1 |
| chr9_+_14551758 | 2.99 |
ENSRNOT00000017157
|
Nfya
|
nuclear transcription factor Y subunit alpha |
| chr10_-_59112788 | 2.95 |
ENSRNOT00000041886
|
Spns3
|
SPNS sphingolipid transporter 3 |
| chr12_+_12374790 | 2.84 |
ENSRNOT00000001347
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chr4_-_155051429 | 2.82 |
ENSRNOT00000020094
|
Klrg1
|
killer cell lectin like receptor G1 |
| chr8_-_65587427 | 2.80 |
ENSRNOT00000016491
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr2_+_4942775 | 2.69 |
ENSRNOT00000093548
ENSRNOT00000093741 |
Fam172a
|
family with sequence similarity 172, member A |
| chr2_+_212435728 | 2.69 |
ENSRNOT00000046960
|
AABR07012868.1
|
|
| chr7_-_119716238 | 2.66 |
ENSRNOT00000075678
|
Il2rb
|
interleukin 2 receptor subunit beta |
| chr16_-_85305782 | 2.66 |
ENSRNOT00000067511
ENSRNOT00000076737 |
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr20_+_27954433 | 2.61 |
ENSRNOT00000064288
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr4_-_119327822 | 2.61 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr17_-_46794845 | 2.58 |
ENSRNOT00000077910
ENSRNOT00000090663 ENSRNOT00000078331 |
Elmo1
|
engulfment and cell motility 1 |
| chr15_-_57805184 | 2.57 |
ENSRNOT00000000168
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr14_+_78939903 | 2.56 |
ENSRNOT00000078562
ENSRNOT00000088221 |
Man2b2
Mrfap1
|
mannosidase, alpha, class 2B, member 2 Morf4 family associated protein 1 |
| chr11_+_24263281 | 2.54 |
ENSRNOT00000086946
|
Gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr8_+_77107536 | 2.53 |
ENSRNOT00000083255
|
Adam10
|
ADAM metallopeptidase domain 10 |
| chr7_+_118895405 | 2.52 |
ENSRNOT00000092095
|
RGD1309808
|
similar to apolipoprotein L2; apolipoprotein L-II |
| chr8_-_28075514 | 2.48 |
ENSRNOT00000072851
|
Vps26b
|
VPS26 retromer complex component B |
| chr9_+_65614142 | 2.47 |
ENSRNOT00000016613
|
Casp8
|
caspase 8 |
| chr8_+_45797315 | 2.42 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr1_+_215609645 | 2.40 |
ENSRNOT00000076140
ENSRNOT00000027487 |
Tnni2
|
troponin I2, fast skeletal type |
| chr11_-_51202703 | 2.36 |
ENSRNOT00000002719
ENSRNOT00000077220 |
Cblb
|
Cbl proto-oncogene B |
| chr20_+_28815039 | 2.34 |
ENSRNOT00000075800
|
Sowahc
|
sosondowah ankyrin repeat domain family member C |
| chr10_+_57268375 | 2.28 |
ENSRNOT00000005242
|
Rnf167
|
ring finger protein 167 |
| chr13_-_47979797 | 2.26 |
ENSRNOT00000080035
|
Rassf5
|
Ras association domain family member 5 |
| chr3_+_28627084 | 2.26 |
ENSRNOT00000049884
|
Arhgap15
|
Rho GTPase activating protein 15 |
| chr5_-_79222687 | 2.24 |
ENSRNOT00000010516
|
Akna
|
AT-hook transcription factor |
| chr11_+_84745904 | 2.23 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr3_-_5976244 | 2.23 |
ENSRNOT00000009893
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr19_+_43392085 | 2.21 |
ENSRNOT00000023854
|
Sf3b3
|
splicing factor 3b, subunit 3 |
| chrX_+_15155230 | 2.12 |
ENSRNOT00000073289
ENSRNOT00000051439 |
Was
|
Wiskott-Aldrich syndrome |
| chr6_+_136358057 | 2.09 |
ENSRNOT00000092741
|
Klc1
|
kinesin light chain 1 |
| chr9_-_116222374 | 2.07 |
ENSRNOT00000090111
ENSRNOT00000067900 |
Arhgap28
|
Rho GTPase activating protein 28 |
| chr3_-_3476215 | 2.07 |
ENSRNOT00000024352
|
Ubac1
|
UBA domain containing 1 |
| chr1_-_265798167 | 2.04 |
ENSRNOT00000079483
|
Ldb1
|
LIM domain binding 1 |
| chr15_-_33766438 | 2.01 |
ENSRNOT00000033977
|
Ap1g2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chr5_+_60250546 | 2.01 |
ENSRNOT00000017707
|
Zcchc7
|
zinc finger CCHC-type containing 7 |
| chr1_+_222250995 | 2.01 |
ENSRNOT00000031458
|
Trpt1
|
tRNA phosphotransferase 1 |
| chr1_-_175420106 | 2.01 |
ENSRNOT00000013126
ENSRNOT00000077125 |
Sbf2
|
SET binding factor 2 |
| chr9_+_81880177 | 2.00 |
ENSRNOT00000022839
|
Stk36
|
serine/threonine kinase 36 |
| chr5_+_156668712 | 1.99 |
ENSRNOT00000067228
|
Ddost
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase non-catalytic subunit |
| chr10_-_47164596 | 1.93 |
ENSRNOT00000075751
|
LOC100911674
|
protein GTLF3B-like |
| chr16_+_20824864 | 1.90 |
ENSRNOT00000092293
ENSRNOT00000092446 |
Upf1
|
UPF1, RNA helicase and ATPase |
| chr10_+_55492404 | 1.89 |
ENSRNOT00000005588
ENSRNOT00000078038 |
Rpl26
|
ribosomal protein L26 |
| chr8_+_116307141 | 1.88 |
ENSRNOT00000037375
|
Rassf1
|
Ras association domain family member 1 |
| chr1_+_197999037 | 1.88 |
ENSRNOT00000091065
|
Apobr
|
apolipoprotein B receptor |
| chr4_-_100099517 | 1.86 |
ENSRNOT00000014277
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr15_+_34352914 | 1.85 |
ENSRNOT00000067150
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr1_+_201256910 | 1.83 |
ENSRNOT00000090143
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr4_+_140247313 | 1.82 |
ENSRNOT00000040255
ENSRNOT00000064025 ENSRNOT00000041130 ENSRNOT00000043646 |
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr10_+_17421075 | 1.81 |
ENSRNOT00000047011
|
Stk10
|
serine/threonine kinase 10 |
| chr20_-_4818568 | 1.79 |
ENSRNOT00000001115
|
Ddx39b
|
DExD-box helicase 39B |
| chr12_+_13716596 | 1.79 |
ENSRNOT00000080216
|
Actb
|
actin, beta |
| chr2_+_187993758 | 1.77 |
ENSRNOT00000027182
|
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr16_+_20825044 | 1.75 |
ENSRNOT00000027268
|
Upf1
|
UPF1, RNA helicase and ATPase |
| chr7_-_120027026 | 1.74 |
ENSRNOT00000011215
|
Card10
|
caspase recruitment domain family, member 10 |
| chr9_+_46840992 | 1.73 |
ENSRNOT00000019415
|
Il1r2
|
interleukin 1 receptor type 2 |
| chr15_+_57340579 | 1.73 |
ENSRNOT00000015467
|
Zc3h13
|
zinc finger CCCH type containing 13 |
| chr20_+_5125349 | 1.71 |
ENSRNOT00000085598
ENSRNOT00000001129 |
Bag6
|
BCL2-associated athanogene 6 |
| chr10_-_103481153 | 1.70 |
ENSRNOT00000035639
|
Cd300lb
|
CD300 molecule-like family member b |
| chr15_+_33108671 | 1.69 |
ENSRNOT00000015468
|
Lrp10
|
LDL receptor related protein 10 |
| chr12_+_11179329 | 1.62 |
ENSRNOT00000001302
|
Zfp394
|
zinc finger protein 394 |
| chr8_+_61080103 | 1.58 |
ENSRNOT00000022976
|
Hmg20a
|
high mobility group 20A |
| chr19_+_37568113 | 1.57 |
ENSRNOT00000023710
|
Fam65a
|
family with sequence similarity 65, member A |
| chr16_+_74177215 | 1.57 |
ENSRNOT00000025851
|
Ikbkb
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr8_-_6076598 | 1.56 |
ENSRNOT00000090774
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr2_-_33025271 | 1.55 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_198199622 | 1.53 |
ENSRNOT00000026688
|
Gdpd3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr1_-_101118825 | 1.52 |
ENSRNOT00000066328
|
Rps11
|
ribosomal protein S11 |
| chr10_-_105795958 | 1.50 |
ENSRNOT00000067739
|
Srsf2
|
serine and arginine rich splicing factor 2 |
| chr3_-_111037166 | 1.50 |
ENSRNOT00000017070
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr5_+_104941066 | 1.47 |
ENSRNOT00000009225
|
Rraga
|
Ras-related GTP binding A |
| chr9_+_94880053 | 1.47 |
ENSRNOT00000024445
|
Atg16l1
|
autophagy related 16-like 1 |
| chr20_+_9948908 | 1.46 |
ENSRNOT00000001541
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr4_+_157523320 | 1.46 |
ENSRNOT00000023192
|
Zfp384
|
zinc finger protein 384 |
| chr9_+_94879745 | 1.46 |
ENSRNOT00000080482
|
Atg16l1
|
autophagy related 16-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ets2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 3.2 | 31.6 | GO:0090500 | dorsal aorta morphogenesis(GO:0035912) endocardial cushion to mesenchymal transition(GO:0090500) |
| 2.7 | 8.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 2.5 | 7.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 2.5 | 7.5 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 2.3 | 6.8 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 1.9 | 9.5 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 1.7 | 15.4 | GO:0008228 | opsonization(GO:0008228) |
| 1.7 | 6.8 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 1.5 | 10.7 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 1.5 | 4.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 1.4 | 4.3 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 1.4 | 4.3 | GO:0048850 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) hypophysis morphogenesis(GO:0048850) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) melanocyte proliferation(GO:0097325) |
| 1.4 | 8.5 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 1.4 | 4.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 1.3 | 9.3 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 1.2 | 3.7 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 1.2 | 14.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 1.2 | 8.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 1.2 | 6.9 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 1.1 | 21.7 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 1.1 | 6.6 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 1.1 | 5.4 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 1.0 | 4.2 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 1.0 | 4.9 | GO:0042117 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) monocyte activation(GO:0042117) multi-organism membrane fusion(GO:0044800) |
| 0.9 | 5.2 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.8 | 8.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.8 | 4.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.8 | 6.9 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.7 | 10.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.7 | 5.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.7 | 4.3 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.7 | 7.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.7 | 3.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.7 | 4.0 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.6 | 1.9 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.6 | 8.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.6 | 1.7 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.5 | 2.7 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.5 | 2.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.5 | 4.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.5 | 7.3 | GO:0051639 | actin filament network formation(GO:0051639) actin crosslink formation(GO:0051764) |
| 0.5 | 2.9 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.5 | 1.4 | GO:0002332 | transitional stage B cell differentiation(GO:0002332) transitional one stage B cell differentiation(GO:0002333) positive regulation of actin filament binding(GO:1904531) positive regulation of actin binding(GO:1904618) activation of protein kinase C activity(GO:1990051) |
| 0.5 | 5.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 4.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.4 | 5.8 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.4 | 8.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.4 | 5.8 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.4 | 1.8 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.4 | 4.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.4 | 4.7 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.4 | 2.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.4 | 1.2 | GO:1902544 | positive regulation of endodeoxyribonuclease activity(GO:0032079) regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.4 | 1.2 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.4 | 3.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 2.6 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.4 | 2.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.4 | 1.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.4 | 13.4 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.4 | 5.7 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.3 | 1.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.3 | 3.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 5.1 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.3 | 6.0 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.3 | 0.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 3.6 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 5.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.3 | 3.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.3 | 12.3 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.3 | 0.8 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.3 | 1.8 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.3 | 1.3 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.3 | 1.6 | GO:1903347 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.3 | 4.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 | 6.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 0.9 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 2.4 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 | 14.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.2 | 2.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.7 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 0.2 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.2 | 7.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 0.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of neutrophil activation(GO:1902564) |
| 0.2 | 3.6 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.2 | 6.7 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 8.9 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.2 | 3.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.2 | 2.9 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 0.4 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.2 | 2.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 7.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 1.0 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 2.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.2 | 4.8 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.2 | 0.7 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.2 | 2.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.2 | 3.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.2 | 3.8 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 1.4 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 5.6 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 0.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.8 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 3.2 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.1 | 0.7 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 3.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.4 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 18.9 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 1.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.9 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 1.8 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 4.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 2.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.3 | GO:0035811 | negative regulation of urine volume(GO:0035811) regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 1.7 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.1 | 1.8 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.1 | 3.9 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.1 | 0.3 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 1.9 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 4.1 | GO:0048265 | response to pain(GO:0048265) |
| 0.1 | 1.5 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.5 | GO:0046070 | dGTP metabolic process(GO:0046070) |
| 0.1 | 1.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.5 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.1 | 0.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.9 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 3.7 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.3 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.1 | 3.0 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.1 | GO:0071454 | cellular response to anoxia(GO:0071454) |
| 0.1 | 0.9 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 0.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 3.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.7 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 2.5 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 5.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 1.5 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.1 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.9 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 1.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.8 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 1.1 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 1.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.6 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 1.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 4.6 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.4 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.4 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 2.5 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 1.2 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.8 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 3.3 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 4.2 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 2.9 | 11.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 2.5 | 7.5 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 1.7 | 5.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.6 | 13.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.5 | 9.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.4 | 6.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.1 | 10.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 1.0 | 4.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.0 | 5.7 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.9 | 25.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.8 | 2.5 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.8 | 3.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.7 | 3.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.6 | 7.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 1.8 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.6 | 1.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.5 | 1.8 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.4 | 3.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.4 | 15.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.4 | 5.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 13.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 6.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.3 | 2.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 6.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 4.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.3 | 1.5 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.3 | 1.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 4.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 6.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 1.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 1.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 27.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 1.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 2.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 1.2 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.2 | 1.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 1.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 4.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 4.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 1.8 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.7 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 2.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 4.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 3.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.2 | 0.6 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 4.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 4.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 17.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 3.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.9 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 2.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 2.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 23.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 5.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 0.5 | GO:0044294 | SCAR complex(GO:0031209) dendritic growth cone(GO:0044294) |
| 0.1 | 3.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 11.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 21.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.1 | 7.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 3.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 5.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 21.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 9.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 4.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 2.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0000940 | condensed nuclear chromosome kinetochore(GO:0000778) condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 2.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 9.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 3.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 19.3 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 22.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) BMP receptor activity(GO:0098821) |
| 2.2 | 8.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 2.2 | 17.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.8 | 9.2 | GO:0005534 | galactose binding(GO:0005534) |
| 1.8 | 11.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.7 | 6.8 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 1.7 | 13.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.6 | 4.9 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 1.4 | 4.3 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 1.4 | 4.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.3 | 5.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.2 | 9.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 1.2 | 4.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.1 | 7.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.0 | 7.8 | GO:0038085 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 0.9 | 4.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.9 | 6.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.8 | 7.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.8 | 5.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.7 | 4.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.6 | 3.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.6 | 2.5 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.6 | 1.8 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.6 | 4.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.6 | 2.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.6 | 3.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.6 | 5.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 4.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 1.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.5 | 7.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 6.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 2.9 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.5 | 1.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.4 | 9.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.4 | 4.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.4 | 1.2 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.4 | 3.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.4 | 1.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.4 | 8.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 3.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 9.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.3 | 1.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.3 | 2.7 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.3 | 5.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 14.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 6.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.3 | 15.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.3 | 6.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.3 | 3.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 4.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 1.2 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.2 | 2.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 5.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 8.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 5.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.8 | GO:0030621 | U6 snRNA binding(GO:0017070) U4 snRNA binding(GO:0030621) |
| 0.2 | 1.7 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.2 | 12.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 3.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 1.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 1.0 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.2 | 4.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 5.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.5 | GO:0019961 | interferon binding(GO:0019961) |
| 0.2 | 48.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 1.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 1.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 4.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 0.8 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 0.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 3.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 2.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 4.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 6.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 3.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 3.2 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 9.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 6.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 1.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 2.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 3.3 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.1 | 1.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 22.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 1.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 2.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 6.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 3.0 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 2.2 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 5.6 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 7.1 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 10.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 3.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.0 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.9 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 4.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 34.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 1.0 | 27.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.9 | 31.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.7 | 7.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 9.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.4 | 17.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.4 | 7.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.3 | 20.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 8.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.3 | 16.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.3 | 20.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.2 | 4.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 35.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 27.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 1.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 3.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 4.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 5.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.8 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 1.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 16.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.9 | 8.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.8 | 28.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.8 | 7.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.7 | 10.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.6 | 13.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.6 | 6.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.5 | 4.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.5 | 7.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.5 | 4.3 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.5 | 6.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.5 | 8.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 7.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.4 | 6.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 4.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 52.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.4 | 2.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.3 | 6.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 4.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.3 | 4.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 4.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 3.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 3.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 5.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 5.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 2.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 17.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 7.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 1.9 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.2 | 1.3 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.1 | 3.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 4.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.8 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 2.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 4.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 4.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 3.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 4.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |