Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
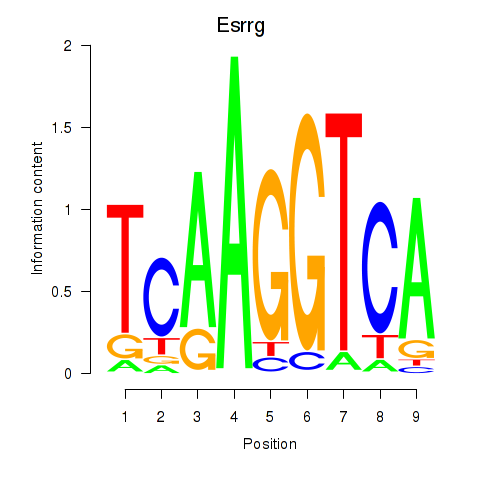
Results for Esrrg
Z-value: 0.69
Transcription factors associated with Esrrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esrrg
|
ENSRNOG00000002593 | estrogen-related receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esrrg | rn6_v1_chr13_+_106463368_106463368 | 0.44 | 2.5e-16 | Click! |
Activity profile of Esrrg motif
Sorted Z-values of Esrrg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_126080698 | 22.83 |
ENSRNOT00000008739
|
Bsnd
|
barttin CLCNK type accessory beta subunit |
| chr13_-_85622314 | 21.04 |
ENSRNOT00000005719
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr3_+_151126591 | 20.46 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr5_-_126911520 | 19.58 |
ENSRNOT00000091521
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr8_+_71167305 | 17.94 |
ENSRNOT00000021337
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr3_-_60813869 | 12.99 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr7_+_2504695 | 11.63 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr9_+_61692154 | 11.22 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr4_-_82209933 | 10.91 |
ENSRNOT00000091106
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr10_+_61432819 | 10.56 |
ENSRNOT00000003687
ENSRNOT00000092478 |
Cluh
|
clustered mitochondria homolog |
| chr4_-_82295470 | 10.38 |
ENSRNOT00000091073
|
Hoxa10
|
homeobox A10 |
| chr7_-_120518653 | 10.10 |
ENSRNOT00000016362
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr19_-_10653800 | 9.59 |
ENSRNOT00000022128
|
Cx3cl1
|
C-X3-C motif chemokine ligand 1 |
| chr7_-_81592206 | 8.80 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr9_-_61690956 | 8.70 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr13_-_93677377 | 8.69 |
ENSRNOT00000004917
|
Fh
|
fumarate hydratase |
| chrX_-_40086870 | 8.60 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr14_+_17210733 | 8.48 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr4_-_157679962 | 7.97 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr18_+_2416871 | 7.97 |
ENSRNOT00000081399
|
Gata6
|
GATA binding protein 6 |
| chr15_+_61069581 | 7.90 |
ENSRNOT00000084333
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr12_-_48254822 | 7.88 |
ENSRNOT00000056865
|
Ung
|
uracil-DNA glycosylase |
| chr8_-_55037604 | 7.81 |
ENSRNOT00000059169
|
Sdhd
|
succinate dehydrogenase complex subunit D |
| chr8_+_39878955 | 7.70 |
ENSRNOT00000060300
|
LOC100911068
|
roundabout homolog 4-like |
| chr18_+_2416552 | 7.68 |
ENSRNOT00000030726
|
Gata6
|
GATA binding protein 6 |
| chr18_+_74156553 | 7.61 |
ENSRNOT00000022892
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr10_+_10761477 | 6.98 |
ENSRNOT00000004176
|
Rogdi
|
rogdi homolog |
| chr6_+_128750795 | 6.76 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr1_+_190555177 | 6.74 |
ENSRNOT00000021514
|
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chr4_+_51614676 | 6.74 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr1_+_261158261 | 6.67 |
ENSRNOT00000071965
|
Pgam1
|
phosphoglycerate mutase 1 |
| chr7_+_117409576 | 6.43 |
ENSRNOT00000017067
|
Cyc1
|
cytochrome c-1 |
| chr8_-_39093277 | 6.03 |
ENSRNOT00000043344
|
LOC100911068
|
roundabout homolog 4-like |
| chr7_-_80796670 | 6.02 |
ENSRNOT00000010539
|
Abra
|
actin-binding Rho activating protein |
| chr1_-_263269762 | 6.00 |
ENSRNOT00000022309
|
Got1
|
glutamic-oxaloacetic transaminase 1 |
| chr18_-_27749235 | 5.77 |
ENSRNOT00000026696
|
Hspa9
|
heat shock protein family A member 9 |
| chr5_+_159484370 | 5.66 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr5_-_147412705 | 5.43 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr10_-_71849293 | 5.30 |
ENSRNOT00000003799
|
Lhx1
|
LIM homeobox 1 |
| chr8_+_106317124 | 5.01 |
ENSRNOT00000018411
|
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr2_+_186980793 | 4.98 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr5_-_56576676 | 4.96 |
ENSRNOT00000029712
|
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr2_-_47096961 | 4.94 |
ENSRNOT00000077401
|
Itga2
|
integrin alpha 2 |
| chr2_+_186980992 | 4.89 |
ENSRNOT00000020717
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr10_-_11085142 | 4.76 |
ENSRNOT00000081596
ENSRNOT00000005479 ENSRNOT00000085230 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr6_-_4520604 | 4.73 |
ENSRNOT00000042230
ENSRNOT00000043870 ENSRNOT00000070918 ENSRNOT00000046246 ENSRNOT00000052367 ENSRNOT00000042251 |
Slc8a1
|
solute carrier family 8 member A1 |
| chr12_-_30566032 | 4.43 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr16_-_69242028 | 4.05 |
ENSRNOT00000019002
|
Zfp703
|
zinc finger protein 703 |
| chr9_+_43259709 | 3.97 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr4_+_115046693 | 3.84 |
ENSRNOT00000031583
|
Bola3
|
bolA family member 3 |
| chr8_+_59164572 | 3.81 |
ENSRNOT00000015102
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr4_-_157331905 | 3.66 |
ENSRNOT00000020647
|
Tpi1
|
triosephosphate isomerase 1 |
| chr9_+_20213776 | 3.59 |
ENSRNOT00000071439
|
LOC100911515
|
triosephosphate isomerase-like |
| chr16_+_1979191 | 3.58 |
ENSRNOT00000014382
|
Ppif
|
peptidylprolyl isomerase F |
| chr6_+_2216623 | 3.43 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr7_-_117259141 | 3.32 |
ENSRNOT00000040762
|
Plec
|
plectin |
| chr7_+_129860327 | 3.26 |
ENSRNOT00000043461
|
Pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr2_+_252305874 | 3.26 |
ENSRNOT00000020972
|
Ctbs
|
chitobiase |
| chrX_+_158835811 | 3.13 |
ENSRNOT00000071888
ENSRNOT00000080110 |
Ints6l
|
integrator complex subunit 6 like |
| chr7_-_12424367 | 3.08 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr6_+_64252970 | 2.88 |
ENSRNOT00000093700
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr5_+_173640780 | 2.87 |
ENSRNOT00000027476
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr1_+_199555722 | 2.62 |
ENSRNOT00000054983
|
Itgax
|
integrin subunit alpha X |
| chr1_-_78968329 | 2.59 |
ENSRNOT00000023078
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr1_-_263431290 | 2.56 |
ENSRNOT00000022633
|
Slc25a28
|
solute carrier family 25 member 28 |
| chr14_-_114583122 | 2.55 |
ENSRNOT00000084595
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr10_-_20818128 | 2.50 |
ENSRNOT00000011061
|
Wwc1
|
WW and C2 domain containing 1 |
| chr8_-_68312909 | 2.42 |
ENSRNOT00000066106
|
RGD1309779
|
similar to ENSANGP00000021391 |
| chr5_-_100647727 | 2.36 |
ENSRNOT00000067435
|
Nfib
|
nuclear factor I/B |
| chr15_+_61662264 | 2.36 |
ENSRNOT00000072969
|
Mtrf1
|
mitochondrial translation release factor 1 |
| chr10_+_109533487 | 2.32 |
ENSRNOT00000054975
|
Fscn2
|
fascin actin-bundling protein 2, retinal |
| chr5_+_118743632 | 2.04 |
ENSRNOT00000013785
|
Pgm1
|
phosphoglucomutase 1 |
| chr12_-_24365324 | 2.03 |
ENSRNOT00000032250
|
Trim50
|
tripartite motif-containing 50 |
| chr12_+_47024442 | 1.89 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chrX_+_159427745 | 1.88 |
ENSRNOT00000078226
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr3_+_2727254 | 1.78 |
ENSRNOT00000034089
|
Fbxw5
|
F-box and WD repeat domain containing 5 |
| chr18_-_63185510 | 1.76 |
ENSRNOT00000024632
|
Afg3l2
|
AFG3 like matrix AAA peptidase subunit 2 |
| chr4_-_99746560 | 1.75 |
ENSRNOT00000012021
|
Mrpl35
|
mitochondrial ribosomal protein L35 |
| chr1_+_128637049 | 1.73 |
ENSRNOT00000018639
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr6_-_107678156 | 1.66 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr5_-_59085676 | 1.60 |
ENSRNOT00000068105
|
Msmp
|
microseminoprotein, prostate associated |
| chr3_+_147609095 | 1.56 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr7_+_13698647 | 1.53 |
ENSRNOT00000067342
|
Olr1086
|
olfactory receptor 1086 |
| chr7_+_11521998 | 1.51 |
ENSRNOT00000026960
|
Sgta
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr9_-_15628881 | 1.38 |
ENSRNOT00000077318
ENSRNOT00000020950 |
Guca1b
|
guanylate cyclase activator 1B |
| chr1_-_221041401 | 1.24 |
ENSRNOT00000064136
|
Pcnx3
|
pecanex homolog 3 (Drosophila) |
| chr8_-_47404010 | 1.21 |
ENSRNOT00000038647
|
Tmem136
|
transmembrane protein 136 |
| chr18_+_402295 | 1.19 |
ENSRNOT00000033618
|
Fundc2
|
FUN14 domain containing 2 |
| chr3_+_11607225 | 1.14 |
ENSRNOT00000072685
|
St6galnac4
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr5_+_50381244 | 1.03 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr1_+_255842783 | 1.01 |
ENSRNOT00000023562
|
March5
|
membrane associated ring-CH-type finger 5 |
| chr17_-_84247038 | 0.99 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr10_-_88356538 | 0.93 |
ENSRNOT00000022430
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr8_+_55037750 | 0.93 |
ENSRNOT00000013188
|
Timm8b
|
translocase of inner mitochondrial membrane 8 homolog B |
| chr5_-_75319765 | 0.92 |
ENSRNOT00000085698
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr10_+_67559385 | 0.92 |
ENSRNOT00000082685
ENSRNOT00000006218 ENSRNOT00000079696 |
Rhot1
|
ras homolog family member T1 |
| chr3_-_133232322 | 0.80 |
ENSRNOT00000034487
|
Esf1
|
ESF1 nucleolar pre-rRNA processing protein homolog |
| chr2_-_24923128 | 0.79 |
ENSRNOT00000044087
|
Pde8b
|
phosphodiesterase 8B |
| chr10_-_88357025 | 0.70 |
ENSRNOT00000080006
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr3_+_8534440 | 0.69 |
ENSRNOT00000045827
ENSRNOT00000082672 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr15_-_3544685 | 0.67 |
ENSRNOT00000015179
ENSRNOT00000085126 |
Vcl
|
vinculin |
| chr2_+_38230757 | 0.63 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr1_+_145770135 | 0.62 |
ENSRNOT00000015320
|
Stard5
|
StAR-related lipid transfer domain containing 5 |
| chr6_+_55085313 | 0.56 |
ENSRNOT00000005458
|
AABR07063901.1
|
|
| chr5_-_136002900 | 0.31 |
ENSRNOT00000025197
|
Plk3
|
polo-like kinase 3 |
| chr6_-_76552559 | 0.17 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr6_-_93562314 | 0.15 |
ENSRNOT00000010871
ENSRNOT00000088790 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr8_+_12160067 | 0.12 |
ENSRNOT00000036338
|
Maml2
|
mastermind-like transcriptional coactivator 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Esrrg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.6 | GO:0032912 | endodermal cell fate determination(GO:0007493) negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 4.9 | 14.7 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 2.9 | 8.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 2.9 | 11.6 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 2.9 | 8.7 | GO:0045041 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) protein import into mitochondrial intermembrane space(GO:0045041) |
| 2.8 | 8.5 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 2.7 | 8.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 2.6 | 7.8 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 2.6 | 10.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 2.3 | 6.8 | GO:0009249 | protein lipoylation(GO:0009249) |
| 2.0 | 7.9 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 1.9 | 5.8 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 1.7 | 5.0 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 1.6 | 9.6 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 1.3 | 5.3 | GO:0060066 | metanephric part of ureteric bud development(GO:0035502) oviduct development(GO:0060066) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 1.3 | 17.9 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 1.2 | 3.6 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 1.0 | 6.7 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 1.0 | 6.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.0 | 4.8 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.9 | 20.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.8 | 4.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.8 | 2.4 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.7 | 3.7 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.7 | 2.9 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.7 | 4.9 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.7 | 10.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.7 | 11.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.7 | 3.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.6 | 3.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.6 | 1.8 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.6 | 10.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.5 | 1.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.4 | 2.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 5.7 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.3 | 4.0 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 1.0 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.3 | 10.4 | GO:0060065 | uterus development(GO:0060065) |
| 0.3 | 2.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.3 | 20.4 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.3 | 3.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.3 | 2.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 3.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 2.6 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.2 | 21.0 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.2 | 6.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 3.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 1.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 17.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 2.9 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 2.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 5.0 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 0.3 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 1.0 | GO:0006590 | thyroid hormone generation(GO:0006590) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 2.6 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 3.1 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.6 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 8.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 7.0 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 0.9 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 1.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 1.0 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 2.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 2.6 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 8.6 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.7 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.5 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 2.4 | 19.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 2.3 | 20.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.7 | 8.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.7 | 8.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.6 | 4.9 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 1.3 | 3.8 | GO:0045242 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 1.2 | 13.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 1.0 | 10.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 1.0 | 13.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.0 | 8.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.7 | 8.7 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.6 | 1.8 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.5 | 4.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.4 | 3.6 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.4 | 5.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 3.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 2.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 2.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 3.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 5.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 5.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 43.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 2.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 8.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 19.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 2.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 4.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 6.4 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 8.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 12.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 1.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 26.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 8.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 9.3 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 11.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.3 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.5 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 3.8 | 19.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 2.4 | 19.6 | GO:0008430 | selenium binding(GO:0008430) |
| 2.4 | 7.3 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 2.0 | 6.0 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 2.0 | 8.0 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.7 | 8.7 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.7 | 8.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.7 | 5.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 1.7 | 6.7 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.3 | 7.9 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 1.2 | 22.8 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 1.1 | 21.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 1.0 | 4.9 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 1.0 | 3.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.7 | 13.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.7 | 4.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.6 | 2.4 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.5 | 3.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.5 | 1.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.4 | 15.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.4 | 2.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 2.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.4 | 1.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 19.1 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.3 | 9.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.3 | 2.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 21.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 8.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.2 | 5.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 28.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 15.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 2.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 4.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 8.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 6.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 10.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 8.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 5.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 4.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.6 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 10.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 7.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.3 | 15.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 9.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.2 | 8.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 8.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 2.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 8.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 9.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 26.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 1.3 | 20.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 1.2 | 19.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.7 | 21.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.6 | 24.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 21.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.4 | 8.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.4 | 18.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 4.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 17.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 9.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 4.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 3.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 15.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.9 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 2.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 5.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 6.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |