Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
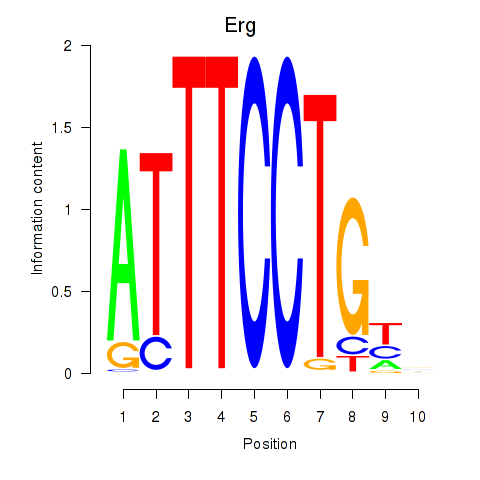
Results for Erg
Z-value: 2.20
Transcription factors associated with Erg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Erg
|
ENSRNOG00000001652 | ERG, ETS transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Erg | rn6_v1_chr11_-_35749464_35749594 | 0.71 | 1.3e-50 | Click! |
Activity profile of Erg motif
Sorted Z-values of Erg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_11077411 | 126.34 |
ENSRNOT00000007117
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr13_+_47602692 | 96.83 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr13_+_89774764 | 91.86 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr11_+_84745904 | 84.06 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr14_-_43992587 | 78.69 |
ENSRNOT00000003425
|
Rhoh
|
ras homolog family member H |
| chr11_-_66759402 | 76.47 |
ENSRNOT00000003326
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr4_-_163227242 | 74.36 |
ENSRNOT00000091552
ENSRNOT00000077793 |
Clec7a
|
C-type lectin domain family 7, member A |
| chr2_-_190100276 | 74.08 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chrX_+_134979646 | 71.11 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr7_-_107392972 | 70.73 |
ENSRNOT00000093425
|
Tmem71
|
transmembrane protein 71 |
| chr1_+_198744050 | 70.09 |
ENSRNOT00000024404
|
Itgal
|
integrin subunit alpha L |
| chr13_+_50164563 | 68.90 |
ENSRNOT00000029533
|
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr7_+_142776252 | 68.77 |
ENSRNOT00000008673
|
Acvrl1
|
activin A receptor like type 1 |
| chr7_+_142776580 | 67.34 |
ENSRNOT00000081047
|
Acvrl1
|
activin A receptor like type 1 |
| chr11_+_54619129 | 65.22 |
ENSRNOT00000059924
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr1_-_227441442 | 65.14 |
ENSRNOT00000028433
|
Ms4a1
|
membrane spanning 4-domains A1 |
| chr16_-_19918644 | 64.50 |
ENSRNOT00000083345
ENSRNOT00000023926 |
Plvap
|
plasmalemma vesicle associated protein |
| chr1_-_197770669 | 63.76 |
ENSRNOT00000023563
|
Lat
|
linker for activation of T cells |
| chr13_-_55173692 | 61.65 |
ENSRNOT00000064785
ENSRNOT00000029878 ENSRNOT00000029865 ENSRNOT00000060292 ENSRNOT00000000814 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr2_+_183674522 | 61.02 |
ENSRNOT00000014433
|
Tmem154
|
transmembrane protein 154 |
| chr16_-_9430743 | 60.47 |
ENSRNOT00000043811
|
Wdfy4
|
WDFY family member 4 |
| chr2_+_93792601 | 58.18 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr13_-_111917587 | 57.18 |
ENSRNOT00000007649
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr2_+_187218851 | 57.03 |
ENSRNOT00000017798
|
Sh2d2a
|
SH2 domain containing 2A |
| chr20_+_30915213 | 56.76 |
ENSRNOT00000000681
|
Prf1
|
perforin 1 |
| chr1_-_198577226 | 55.65 |
ENSRNOT00000055013
|
Spn
|
sialophorin |
| chr8_-_49308806 | 55.10 |
ENSRNOT00000047291
|
Cd3e
|
CD3e molecule |
| chr1_-_198662610 | 54.39 |
ENSRNOT00000055012
|
Sept1
|
septin 1 |
| chr9_+_2190915 | 53.66 |
ENSRNOT00000077417
|
Satb1
|
SATB homeobox 1 |
| chr5_-_147761983 | 53.09 |
ENSRNOT00000012936
|
Lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr1_-_206282575 | 52.65 |
ENSRNOT00000024974
|
Adam12
|
ADAM metallopeptidase domain 12 |
| chr1_+_81763614 | 52.56 |
ENSRNOT00000027254
|
Cd79a
|
CD79a molecule |
| chr1_-_17378047 | 50.16 |
ENSRNOT00000020102
|
Themis
|
thymocyte selection associated |
| chr11_+_57108956 | 49.53 |
ENSRNOT00000035485
|
Cd96
|
CD96 molecule |
| chr19_+_19395655 | 48.91 |
ENSRNOT00000019130
|
Snx20
|
sorting nexin 20 |
| chr1_+_42121636 | 46.03 |
ENSRNOT00000025616
|
Myct1
|
myc target 1 |
| chrX_+_15155230 | 45.98 |
ENSRNOT00000073289
ENSRNOT00000051439 |
Was
|
Wiskott-Aldrich syndrome |
| chr10_-_59112788 | 45.49 |
ENSRNOT00000041886
|
Spns3
|
SPNS sphingolipid transporter 3 |
| chr6_+_98284170 | 43.94 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr10_+_47930633 | 43.86 |
ENSRNOT00000003515
|
Grap
|
GRB2-related adaptor protein |
| chr11_-_35749464 | 43.10 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr1_+_166141954 | 42.88 |
ENSRNOT00000042524
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr16_+_18716019 | 42.50 |
ENSRNOT00000047870
|
Sftpa1
|
surfactant protein A1 |
| chr1_+_81779380 | 42.26 |
ENSRNOT00000065865
ENSRNOT00000080143 ENSRNOT00000089592 ENSRNOT00000080840 |
Arhgef1
|
Rho guanine nucleotide exchange factor 1 |
| chr15_+_52451161 | 39.72 |
ENSRNOT00000018725
|
Dok2
|
docking protein 2 |
| chr10_+_94566928 | 39.16 |
ENSRNOT00000078446
|
Prr29
|
proline rich 29 |
| chr5_+_144160108 | 38.94 |
ENSRNOT00000064972
|
Eva1b
|
eva-1 homolog B |
| chr5_+_149056078 | 37.77 |
ENSRNOT00000083028
|
Laptm5
|
lysosomal protein transmembrane 5 |
| chr2_-_47281421 | 37.51 |
ENSRNOT00000086114
|
Itga1
|
integrin subunit alpha 1 |
| chr3_+_28627084 | 37.33 |
ENSRNOT00000049884
|
Arhgap15
|
Rho GTPase activating protein 15 |
| chr10_-_5260608 | 37.15 |
ENSRNOT00000003572
|
Ciita
|
class II, major histocompatibility complex, transactivator |
| chrX_+_14578264 | 36.31 |
ENSRNOT00000038994
|
Cybb
|
cytochrome b-245 beta chain |
| chr2_+_206342066 | 36.31 |
ENSRNOT00000026556
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr4_-_78342863 | 36.23 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr1_-_14412807 | 36.09 |
ENSRNOT00000074583
|
Tnfaip3
|
TNF alpha induced protein 3 |
| chr6_-_79306443 | 35.90 |
ENSRNOT00000030706
|
Clec14a
|
C-type lectin domain family 14, member A |
| chr13_+_83996080 | 35.60 |
ENSRNOT00000004403
ENSRNOT00000070958 |
Cd247
|
Cd247 molecule |
| chrX_+_78259409 | 35.44 |
ENSRNOT00000049779
|
RGD1560455
|
similar to RIKEN cDNA A630033H20 gene |
| chr18_-_28535828 | 35.14 |
ENSRNOT00000068386
|
Tmem173
|
transmembrane protein 173 |
| chr13_+_74456487 | 34.85 |
ENSRNOT00000065801
|
Angptl1
|
angiopoietin-like 1 |
| chr8_+_60760078 | 34.23 |
ENSRNOT00000063930
|
Pstpip1
|
proline-serine-threonine phosphatase-interacting protein 1 |
| chr7_-_119716238 | 32.72 |
ENSRNOT00000075678
|
Il2rb
|
interleukin 2 receptor subunit beta |
| chr16_-_74710704 | 32.72 |
ENSRNOT00000016859
|
Thsd1
|
thrombospondin type 1 domain containing 1 |
| chr4_+_90990088 | 32.68 |
ENSRNOT00000030320
|
Mmrn1
|
multimerin 1 |
| chr9_+_65620658 | 32.66 |
ENSRNOT00000084498
|
Casp8
|
caspase 8 |
| chr3_+_93920447 | 32.50 |
ENSRNOT00000012625
|
Lmo2
|
LIM domain only 2 |
| chr2_-_105047984 | 32.04 |
ENSRNOT00000014970
|
Cpa3
|
carboxypeptidase A3 |
| chr9_+_94745217 | 31.95 |
ENSRNOT00000051338
|
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr9_+_2202511 | 31.77 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chr5_+_154522119 | 31.58 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr1_-_56683731 | 31.55 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
| chr4_+_71740532 | 31.32 |
ENSRNOT00000023537
|
Zyx
|
zyxin |
| chr5_-_79222687 | 31.31 |
ENSRNOT00000010516
|
Akna
|
AT-hook transcription factor |
| chr2_-_198002625 | 31.26 |
ENSRNOT00000091888
|
BC028528
|
cDNA sequence BC028528 |
| chr5_-_137321121 | 30.96 |
ENSRNOT00000027414
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr7_+_70614617 | 30.17 |
ENSRNOT00000035382
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr13_+_88606894 | 29.93 |
ENSRNOT00000048692
|
Sh2d1b
|
SH2 domain containing 1B |
| chr13_-_50761306 | 29.65 |
ENSRNOT00000076610
ENSRNOT00000004241 |
Prelp
|
proline and arginine rich end leucine rich repeat protein |
| chr17_+_22984844 | 28.81 |
ENSRNOT00000078310
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr1_-_216918969 | 28.75 |
ENSRNOT00000079454
|
Osbpl5
|
oxysterol binding protein-like 5 |
| chr10_+_75088422 | 28.63 |
ENSRNOT00000081951
|
Mpo
|
myeloperoxidase |
| chr20_+_9948908 | 28.48 |
ENSRNOT00000001541
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr1_-_89474252 | 28.11 |
ENSRNOT00000028597
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr4_-_44136815 | 28.09 |
ENSRNOT00000086810
|
Tfec
|
transcription factor EC |
| chr15_-_34469350 | 27.93 |
ENSRNOT00000067536
|
Adcy4
|
adenylate cyclase 4 |
| chr5_-_59025631 | 27.80 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chr3_-_91370167 | 27.70 |
ENSRNOT00000006291
|
Prr5l
|
proline rich 5 like |
| chr11_+_67082193 | 27.67 |
ENSRNOT00000003129
|
Cd86
|
CD86 molecule |
| chr9_+_52023295 | 27.65 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr11_+_64601029 | 27.24 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr2_+_32820322 | 27.14 |
ENSRNOT00000013768
|
Cd180
|
CD180 molecule |
| chr1_-_199823386 | 26.99 |
ENSRNOT00000027375
|
Rgs10
|
regulator of G-protein signaling 10 |
| chrX_-_77700269 | 26.39 |
ENSRNOT00000092418
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr1_-_89473904 | 26.36 |
ENSRNOT00000089474
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr10_+_35133252 | 26.16 |
ENSRNOT00000051916
|
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr4_-_157263890 | 26.03 |
ENSRNOT00000065416
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chrX_+_1311121 | 26.01 |
ENSRNOT00000038909
|
Cfp
|
complement factor properdin |
| chr10_+_77537340 | 25.94 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chr3_+_93920013 | 25.79 |
ENSRNOT00000083527
|
Lmo2
|
LIM domain only 2 |
| chr14_-_81023682 | 25.79 |
ENSRNOT00000081570
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr3_-_153001309 | 25.77 |
ENSRNOT00000027581
|
Sla2
|
Src-like-adaptor 2 |
| chr1_+_88875375 | 25.68 |
ENSRNOT00000028284
|
Tyrobp
|
Tyro protein tyrosine kinase binding protein |
| chr8_+_104106740 | 25.55 |
ENSRNOT00000015015
|
Tfdp2
|
transcription factor Dp-2 |
| chr10_-_14443010 | 25.51 |
ENSRNOT00000022142
|
Tmem204
|
transmembrane protein 204 |
| chr9_+_67699379 | 25.48 |
ENSRNOT00000091237
ENSRNOT00000088183 |
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr18_+_44716226 | 25.39 |
ENSRNOT00000086431
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr3_+_79918969 | 25.00 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr9_+_42871950 | 24.96 |
ENSRNOT00000089673
|
Arid5a
|
AT-rich interaction domain 5A |
| chr5_+_153507093 | 24.86 |
ENSRNOT00000086650
ENSRNOT00000083645 |
Runx3
|
runt-related transcription factor 3 |
| chr2_+_212257225 | 24.80 |
ENSRNOT00000077883
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chr9_+_46657922 | 24.73 |
ENSRNOT00000019180
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr3_+_110367939 | 24.69 |
ENSRNOT00000010406
|
Bub1b
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr8_+_133210473 | 24.49 |
ENSRNOT00000082774
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr7_+_70612103 | 24.43 |
ENSRNOT00000057833
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr20_+_7788084 | 23.94 |
ENSRNOT00000000597
|
Def6
|
DEF6 guanine nucleotide exchange factor |
| chr16_-_7250924 | 23.88 |
ENSRNOT00000025394
|
Stab1
|
stabilin 1 |
| chr8_+_102304095 | 23.81 |
ENSRNOT00000011358
|
Slc9a9
|
solute carrier family 9 member A9 |
| chr4_+_158088505 | 23.10 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr3_+_4034921 | 23.03 |
ENSRNOT00000026318
|
Egfl7
|
EGF-like-domain, multiple 7 |
| chr11_+_64882288 | 22.95 |
ENSRNOT00000077727
|
Pla1a
|
phospholipase A1 member A |
| chr10_+_56764927 | 22.56 |
ENSRNOT00000025308
|
Clec10a
|
C-type lectin domain family 10, member A |
| chr20_+_9743269 | 21.78 |
ENSRNOT00000001533
ENSRNOT00000083505 |
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr12_+_9034308 | 21.78 |
ENSRNOT00000001248
|
Flt1
|
FMS-related tyrosine kinase 1 |
| chr6_+_12362813 | 21.71 |
ENSRNOT00000022370
|
Ston1
|
stonin 1 |
| chr1_-_24191908 | 21.58 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_+_11679530 | 21.57 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr2_-_206222248 | 21.13 |
ENSRNOT00000026075
|
Olfml3
|
olfactomedin-like 3 |
| chr15_-_42898150 | 20.69 |
ENSRNOT00000030036
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chrX_-_38196060 | 20.36 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr7_-_143852119 | 20.21 |
ENSRNOT00000016801
|
Rarg
|
retinoic acid receptor, gamma |
| chr11_-_60882379 | 20.02 |
ENSRNOT00000002799
|
Cd200r1
|
CD200 receptor 1 |
| chr10_-_106781950 | 19.99 |
ENSRNOT00000077495
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr5_+_122019301 | 19.67 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr2_+_242634399 | 19.55 |
ENSRNOT00000035700
|
Emcn
|
endomucin |
| chr4_+_156253079 | 19.44 |
ENSRNOT00000013536
|
Clec4d
|
C-type lectin domain family 4, member D |
| chr18_-_6082997 | 19.43 |
ENSRNOT00000086335
ENSRNOT00000032454 |
Ss18
|
SS18, nBAF chromatin remodeling complex subunit |
| chr5_-_160158386 | 19.35 |
ENSRNOT00000089345
|
Fblim1
|
filamin binding LIM protein 1 |
| chr5_-_70463546 | 19.34 |
ENSRNOT00000043184
|
AC118841.1
|
|
| chr13_+_52645257 | 19.18 |
ENSRNOT00000012801
|
Lad1
|
ladinin 1 |
| chrX_-_84167717 | 19.02 |
ENSRNOT00000006415
|
Pof1b
|
premature ovarian failure 1B |
| chrX_+_18998532 | 18.99 |
ENSRNOT00000004213
|
AABR07037343.1
|
|
| chr10_+_17421075 | 18.90 |
ENSRNOT00000047011
|
Stk10
|
serine/threonine kinase 10 |
| chr8_-_32280869 | 18.90 |
ENSRNOT00000009139
|
St14
|
suppression of tumorigenicity 14 |
| chr20_-_32133431 | 18.71 |
ENSRNOT00000000443
|
Srgn
|
serglycin |
| chr18_-_24312950 | 18.68 |
ENSRNOT00000051911
|
LOC686074
|
similar to 60S ribosomal protein L35 |
| chr8_-_72204730 | 18.50 |
ENSRNOT00000023810
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr9_-_20195566 | 18.15 |
ENSRNOT00000015223
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr10_-_56300077 | 18.11 |
ENSRNOT00000064401
|
Tnfsf12
|
tumor necrosis factor superfamily member 12 |
| chr15_-_108326907 | 18.01 |
ENSRNOT00000016848
|
Gpr18
|
G protein-coupled receptor 18 |
| chr8_+_22559098 | 17.95 |
ENSRNOT00000041091
|
LOC691141
|
hypothetical protein LOC691141 |
| chr2_-_189573280 | 17.91 |
ENSRNOT00000022897
|
Rps27
|
ribosomal protein S27 |
| chr10_-_40764185 | 17.90 |
ENSRNOT00000017486
|
Sparc
|
secreted protein acidic and cysteine rich |
| chr6_+_50528823 | 17.82 |
ENSRNOT00000008321
|
Lamb1
|
laminin subunit beta 1 |
| chr18_+_56379890 | 17.78 |
ENSRNOT00000078764
|
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr3_-_63568464 | 17.36 |
ENSRNOT00000068494
|
AABR07052585.2
|
|
| chr17_-_78812111 | 16.99 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr4_-_155051429 | 16.80 |
ENSRNOT00000020094
|
Klrg1
|
killer cell lectin like receptor G1 |
| chr9_-_29647903 | 16.78 |
ENSRNOT00000019062
|
Ogfrl1
|
opioid growth factor receptor-like 1 |
| chr5_+_82587420 | 16.31 |
ENSRNOT00000014020
|
Tlr4
|
toll-like receptor 4 |
| chr3_+_140024043 | 16.30 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr4_+_28989115 | 15.97 |
ENSRNOT00000075326
|
Gng11
|
G protein subunit gamma 11 |
| chr1_-_90149991 | 15.77 |
ENSRNOT00000076987
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr4_+_167754525 | 15.76 |
ENSRNOT00000007889
|
Etv6
|
ets variant 6 |
| chr2_+_187988925 | 15.69 |
ENSRNOT00000093033
ENSRNOT00000093016 |
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr15_+_67555835 | 15.68 |
ENSRNOT00000045882
|
Pcdh17
|
protocadherin 17 |
| chr7_+_59326518 | 15.37 |
ENSRNOT00000085231
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr11_-_14304603 | 15.35 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr16_-_21362955 | 15.27 |
ENSRNOT00000039607
|
Gmip
|
Gem-interacting protein |
| chr2_-_149325913 | 14.79 |
ENSRNOT00000036690
|
Gpr171
|
G protein-coupled receptor 171 |
| chr4_+_31387420 | 14.62 |
ENSRNOT00000074048
|
LOC100912034
|
guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-11-like |
| chr10_+_63662986 | 14.39 |
ENSRNOT00000088722
|
Scarf1
|
scavenger receptor class F, member 1 |
| chr5_+_104362971 | 14.38 |
ENSRNOT00000058520
|
Adamtsl1
|
ADAMTS-like 1 |
| chr13_-_108841482 | 14.30 |
ENSRNOT00000004632
ENSRNOT00000086775 |
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr4_-_165317573 | 14.10 |
ENSRNOT00000087529
ENSRNOT00000080017 |
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chrX_+_40460047 | 14.03 |
ENSRNOT00000010970
|
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr10_+_75087892 | 14.03 |
ENSRNOT00000065910
|
Mpo
|
myeloperoxidase |
| chr8_-_132178217 | 13.93 |
ENSRNOT00000087427
|
Zdhhc3
|
zinc finger, DHHC-type containing 3 |
| chr5_+_144031402 | 13.52 |
ENSRNOT00000011694
|
Csf3r
|
colony stimulating factor 3 receptor |
| chr17_-_46794845 | 13.44 |
ENSRNOT00000077910
ENSRNOT00000090663 ENSRNOT00000078331 |
Elmo1
|
engulfment and cell motility 1 |
| chr11_-_51202703 | 13.44 |
ENSRNOT00000002719
ENSRNOT00000077220 |
Cblb
|
Cbl proto-oncogene B |
| chr9_+_69497121 | 13.42 |
ENSRNOT00000042562
|
Nrp2
|
neuropilin 2 |
| chr1_+_214440503 | 13.38 |
ENSRNOT00000025964
|
Cracr2b
|
calcium release activated channel regulator 2B |
| chr14_-_35581031 | 13.16 |
ENSRNOT00000003077
|
Pdgfra
|
platelet derived growth factor receptor alpha |
| chr10_-_39604070 | 13.13 |
ENSRNOT00000032333
|
Csf2
|
colony stimulating factor 2 |
| chr10_-_90912287 | 13.06 |
ENSRNOT00000093570
|
Gjc1
|
gap junction protein, gamma 1 |
| chr10_-_104358253 | 13.02 |
ENSRNOT00000005942
|
Caskin2
|
cask-interacting protein 2 |
| chr12_-_24537313 | 12.76 |
ENSRNOT00000001975
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr11_+_82680253 | 12.63 |
ENSRNOT00000077119
ENSRNOT00000075512 |
Liph
|
lipase H |
| chr11_-_33003021 | 12.38 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr16_-_56900052 | 12.23 |
ENSRNOT00000017339
|
Msr1
|
macrophage scavenger receptor 1 |
| chr5_+_163612518 | 12.15 |
ENSRNOT00000071103
|
LOC103692519
|
60S ribosomal protein L9 pseudogene |
| chr3_-_119135391 | 11.90 |
ENSRNOT00000045443
|
Gabpb1
|
GA binding protein transcription factor, beta subunit 1 |
| chr20_+_5646097 | 11.72 |
ENSRNOT00000090925
|
Itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr17_+_9837402 | 11.56 |
ENSRNOT00000076436
|
Rab24
|
RAB24, member RAS oncogene family |
| chr3_+_151691515 | 11.44 |
ENSRNOT00000052236
|
AC118414.1
|
|
| chr5_-_137238354 | 11.15 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr12_+_13090172 | 11.11 |
ENSRNOT00000092558
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr3_-_11102515 | 10.96 |
ENSRNOT00000035580
|
Fam78a
|
family with sequence similarity 78, member A |
| chr11_-_61499557 | 10.89 |
ENSRNOT00000046208
|
Usf3
|
upstream transcription factor family member 3 |
| chr10_-_32471454 | 10.72 |
ENSRNOT00000003224
|
Sgcd
|
sarcoglycan, delta |
Network of associatons between targets according to the STRING database.
First level regulatory network of Erg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.1 | 72.4 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 20.6 | 61.7 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 20.1 | 200.8 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 19.1 | 76.5 | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 18.6 | 74.4 | GO:0071226 | cellular response to molecule of fungal origin(GO:0071226) |
| 18.6 | 55.7 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 18.5 | 55.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 18.1 | 54.4 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 14.2 | 42.7 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 12.4 | 49.5 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 12.1 | 36.3 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 11.5 | 171.8 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 10.8 | 64.8 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 9.2 | 27.7 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 8.8 | 96.8 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 8.2 | 24.5 | GO:2000464 | positive regulation of astrocyte chemotaxis(GO:2000464) |
| 8.2 | 65.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 8.0 | 31.9 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 7.7 | 30.9 | GO:0072276 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 7.7 | 46.0 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 7.6 | 53.1 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 7.4 | 103.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 7.2 | 50.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 7.0 | 70.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 6.9 | 27.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 6.9 | 20.7 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 6.8 | 94.8 | GO:0002467 | germinal center formation(GO:0002467) |
| 6.7 | 20.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 6.6 | 19.9 | GO:0010070 | zygote asymmetric cell division(GO:0010070) |
| 6.5 | 32.7 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 6.4 | 25.5 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 6.3 | 31.6 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 6.2 | 25.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 6.2 | 18.7 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 6.2 | 37.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 5.9 | 17.8 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 5.9 | 71.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 5.8 | 23.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 5.4 | 21.8 | GO:0034436 | glycoprotein transport(GO:0034436) response to high density lipoprotein particle(GO:0055099) |
| 5.3 | 58.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 5.2 | 25.9 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 4.9 | 24.7 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 4.9 | 19.6 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 4.7 | 18.9 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 4.7 | 42.5 | GO:0008228 | opsonization(GO:0008228) |
| 4.5 | 13.4 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 4.4 | 26.4 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 4.3 | 25.8 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 4.3 | 17.2 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 4.1 | 12.4 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 3.9 | 15.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 3.9 | 50.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 3.8 | 56.8 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 3.6 | 21.8 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 3.6 | 43.5 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 3.6 | 32.0 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 3.5 | 10.5 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 3.5 | 10.5 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 3.4 | 6.9 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 3.4 | 30.6 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 3.3 | 13.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 3.3 | 13.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 3.2 | 19.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 3.2 | 22.6 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 3.2 | 35.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 3.1 | 62.7 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 3.0 | 48.6 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 3.0 | 32.7 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 2.8 | 17.0 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 2.8 | 8.4 | GO:0060939 | myofibroblast differentiation(GO:0036446) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) regulation of myofibroblast differentiation(GO:1904760) |
| 2.7 | 41.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 2.6 | 68.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 2.6 | 10.2 | GO:0090309 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 2.4 | 19.4 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 2.4 | 21.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 2.4 | 7.1 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 2.3 | 14.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 2.3 | 77.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 2.3 | 18.1 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 2.3 | 4.5 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 2.2 | 37.5 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 2.1 | 12.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 2.1 | 17.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 2.1 | 10.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 2.0 | 28.5 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 2.0 | 25.8 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 2.0 | 7.9 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.9 | 5.8 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 1.9 | 5.8 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 1.9 | 24.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 1.9 | 13.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 1.9 | 39.8 | GO:0001945 | lymph vessel development(GO:0001945) |
| 1.9 | 142.5 | GO:0045576 | mast cell activation(GO:0045576) |
| 1.9 | 5.6 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 1.9 | 11.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 1.8 | 23.8 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 1.8 | 8.9 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 1.8 | 14.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 1.7 | 5.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.6 | 26.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 1.6 | 4.9 | GO:2001268 | response to high light intensity(GO:0009644) cellular response to iron(III) ion(GO:0071283) negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 1.6 | 4.8 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 1.6 | 15.7 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 1.5 | 4.5 | GO:0002191 | cap-dependent translational initiation(GO:0002191) positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 1.5 | 19.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 1.5 | 24.9 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 1.4 | 10.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 1.4 | 12.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 1.3 | 4.0 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 1.3 | 35.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 1.2 | 14.4 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 1.1 | 3.3 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) negative regulation of neutrophil degranulation(GO:0043314) negative regulation of neutrophil activation(GO:1902564) |
| 1.1 | 35.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 1.1 | 35.9 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 1.0 | 6.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 1.0 | 9.3 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 1.0 | 25.0 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 1.0 | 13.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 1.0 | 44.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.9 | 7.3 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.9 | 6.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.9 | 31.0 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.8 | 19.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.8 | 5.8 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.8 | 2.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.8 | 3.3 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.8 | 8.7 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.8 | 6.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.8 | 2.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.8 | 23.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.7 | 2.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.7 | 6.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.7 | 9.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.6 | 4.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.6 | 10.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.6 | 17.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.6 | 13.4 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.6 | 35.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.6 | 25.4 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.6 | 1.2 | GO:0060661 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.6 | 9.6 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.6 | 23.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.6 | 6.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.6 | 4.0 | GO:0006013 | mannose metabolic process(GO:0006013) protein deglycosylation(GO:0006517) |
| 0.6 | 9.0 | GO:0030220 | platelet formation(GO:0030220) |
| 0.6 | 6.1 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.6 | 54.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.5 | 10.4 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.5 | 2.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.5 | 31.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.5 | 5.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.5 | 10.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.5 | 1.5 | GO:1903237 | regulation of leukocyte tethering or rolling(GO:1903236) negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.5 | 27.8 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.5 | 5.1 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.4 | 2.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.4 | 13.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.4 | 5.8 | GO:0001553 | luteinization(GO:0001553) |
| 0.4 | 4.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.4 | 28.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.4 | 16.0 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.4 | 13.5 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.4 | 1.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.4 | 2.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.4 | 29.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.4 | 1.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.4 | 8.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.4 | 12.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.3 | 29.6 | GO:0007569 | cell aging(GO:0007569) |
| 0.3 | 143.3 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.3 | 6.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.3 | 5.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.3 | 13.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.3 | 2.5 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.3 | 6.3 | GO:0019068 | virion assembly(GO:0019068) |
| 0.3 | 32.7 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.3 | 6.1 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.3 | 4.5 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.2 | 5.6 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.2 | 10.9 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.2 | 4.1 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 10.6 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.2 | 19.6 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.2 | 1.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 40.6 | GO:0071593 | leukocyte aggregation(GO:0070486) lymphocyte aggregation(GO:0071593) |
| 0.2 | 15.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 1.9 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.2 | 0.8 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 8.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.2 | 21.2 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 1.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.4 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 4.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.3 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 10.3 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.1 | 0.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 10.7 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.1 | 3.7 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 2.5 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.0 | 4.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 5.2 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 3.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.6 | 116.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 13.1 | 52.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 10.9 | 32.7 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 9.5 | 56.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 7.6 | 265.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 7.2 | 65.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 5.6 | 33.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 5.1 | 55.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 4.5 | 17.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 4.5 | 17.8 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-2 complex(GO:0005607) laminin-10 complex(GO:0043259) |
| 4.3 | 34.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 4.1 | 16.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 4.0 | 27.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 3.6 | 54.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 2.7 | 42.7 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 2.4 | 23.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 2.3 | 27.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 2.2 | 8.7 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 2.0 | 27.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 1.9 | 24.7 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 1.9 | 17.0 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 1.8 | 63.0 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 1.8 | 10.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.7 | 5.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 1.7 | 6.7 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 1.6 | 19.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 1.6 | 50.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 1.6 | 14.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 1.5 | 4.5 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 1.4 | 19.6 | GO:0060091 | kinocilium(GO:0060091) |
| 1.4 | 17.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 1.3 | 5.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 1.2 | 130.5 | GO:0016605 | PML body(GO:0016605) |
| 1.2 | 23.6 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 1.2 | 20.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.2 | 338.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 1.1 | 42.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 1.1 | 27.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.0 | 19.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 1.0 | 6.2 | GO:0036396 | MIS complex(GO:0036396) |
| 1.0 | 23.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 1.0 | 15.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.0 | 2.9 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.9 | 23.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.9 | 6.4 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.9 | 19.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.8 | 19.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.8 | 23.3 | GO:0002102 | podosome(GO:0002102) |
| 0.8 | 6.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.8 | 18.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.8 | 64.8 | GO:0005901 | caveola(GO:0005901) |
| 0.7 | 5.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.7 | 89.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.7 | 10.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.7 | 4.8 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.7 | 75.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.7 | 11.9 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.7 | 42.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.7 | 13.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.6 | 5.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.6 | 9.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.6 | 42.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.6 | 228.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.6 | 18.9 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.6 | 31.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.6 | 11.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.5 | 4.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 2.7 | GO:1990393 | 3M complex(GO:1990393) |
| 0.4 | 22.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.4 | 9.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.4 | 31.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 23.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 4.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 18.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.3 | 62.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.3 | 134.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.3 | 32.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.3 | 18.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 5.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 87.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 1.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 6.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 0.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 21.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.2 | 6.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 11.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 38.0 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.1 | 6.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 19.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 2.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 22.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 28.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 6.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 8.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 22.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 3.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 2.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 50.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 2.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.7 | 136.1 | GO:0098821 | activin receptor activity, type I(GO:0016361) BMP receptor activity(GO:0098821) |
| 13.3 | 53.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 12.6 | 126.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 12.3 | 74.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 10.9 | 32.7 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 10.3 | 30.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 9.7 | 87.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 8.2 | 32.7 | GO:0035877 | death effector domain binding(GO:0035877) |
| 8.2 | 24.5 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 8.0 | 31.9 | GO:0051425 | inositol bisphosphate phosphatase activity(GO:0016312) PTB domain binding(GO:0051425) |
| 7.9 | 39.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 7.6 | 45.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 7.5 | 37.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 7.3 | 21.8 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) sterol-transporting ATPase activity(GO:0034041) glycoprotein transporter activity(GO:0034437) |
| 7.0 | 42.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 6.0 | 36.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 5.3 | 26.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 5.1 | 137.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 5.0 | 90.7 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 5.0 | 35.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 4.7 | 14.2 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 4.5 | 13.5 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 4.5 | 35.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 4.3 | 21.6 | GO:0005534 | galactose binding(GO:0005534) |
| 4.3 | 17.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 4.0 | 36.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 3.9 | 50.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 3.8 | 23.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 3.5 | 24.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 3.5 | 10.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 3.3 | 13.1 | GO:0086077 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 2.7 | 21.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 2.6 | 20.8 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 2.5 | 27.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 2.4 | 29.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 2.4 | 17.0 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 2.4 | 23.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 2.4 | 49.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 2.3 | 334.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 2.2 | 48.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 2.1 | 6.4 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 2.1 | 6.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 2.1 | 28.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 2.0 | 10.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 1.9 | 55.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 1.9 | 5.6 | GO:0019961 | interferon binding(GO:0019961) |
| 1.8 | 19.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 1.7 | 72.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 1.7 | 56.8 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 1.7 | 10.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 1.7 | 48.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 1.6 | 19.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 1.6 | 58.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 1.5 | 9.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.5 | 15.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 1.5 | 4.5 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 1.5 | 13.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.4 | 58.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 1.4 | 5.6 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.3 | 19.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 1.3 | 47.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 1.2 | 3.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 1.2 | 7.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 1.2 | 32.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.2 | 44.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 1.1 | 4.5 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.1 | 8.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.1 | 27.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 1.1 | 49.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 1.1 | 64.7 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 1.1 | 25.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 1.0 | 23.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 1.0 | 9.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 1.0 | 42.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 1.0 | 10.9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.9 | 47.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.8 | 74.2 | GO:0019210 | kinase inhibitor activity(GO:0019210) |
| 0.8 | 7.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.8 | 17.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.7 | 28.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.7 | 47.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.7 | 10.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.7 | 31.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.7 | 5.2 | GO:0017070 | U6 snRNA binding(GO:0017070) U4 snRNA binding(GO:0030621) |
| 0.6 | 26.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.6 | 4.8 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.6 | 36.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.6 | 4.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.6 | 9.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.5 | 5.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 43.7 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.5 | 1.6 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.5 | 5.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.4 | 17.0 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.4 | 4.0 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.4 | 8.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.4 | 29.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.4 | 13.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.4 | 6.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.4 | 11.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.4 | 4.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.4 | 91.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.4 | 16.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.4 | 3.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.4 | 44.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.3 | 6.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 82.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.3 | 226.6 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.3 | 98.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.2 | 6.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 18.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.2 | 6.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.2 | 2.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 15.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 2.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 4.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 29.5 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.1 | 38.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 8.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 53.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 2.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 1.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 11.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 115.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 6.6 | 132.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 5.8 | 99.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 5.8 | 379.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 4.9 | 157.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 3.7 | 107.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 3.6 | 39.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 3.3 | 55.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 3.2 | 85.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 3.0 | 32.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 3.0 | 17.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 2.7 | 30.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 2.7 | 171.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 2.3 | 106.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 2.2 | 13.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 2.2 | 43.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 2.1 | 91.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 2.0 | 36.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 2.0 | 31.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.9 | 52.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 1.8 | 31.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 1.7 | 58.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 1.5 | 52.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 1.4 | 74.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 1.4 | 31.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 1.2 | 4.9 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 1.2 | 48.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 1.2 | 17.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.1 | 17.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 1.1 | 24.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 1.1 | 17.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 1.0 | 36.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 1.0 | 160.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 1.0 | 30.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 1.0 | 29.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.9 | 24.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.9 | 13.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.9 | 42.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.9 | 21.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.8 | 43.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.7 | 32.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.7 | 38.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.7 | 24.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.6 | 7.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.6 | 25.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.6 | 2.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.6 | 17.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.6 | 28.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.4 | 5.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.4 | 71.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.4 | 6.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.4 | 6.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.3 | 21.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 10.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 8.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 5.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 35.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.7 | 205.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 5.7 | 114.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 5.2 | 78.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 5.0 | 20.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 4.1 | 32.7 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 3.9 | 58.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 3.6 | 39.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 3.3 | 39.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 3.3 | 98.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 3.3 | 16.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 3.3 | 65.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 3.2 | 139.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 3.2 | 48.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 2.9 | 48.7 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 2.9 | 34.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 2.7 | 65.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 2.3 | 339.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 2.3 | 23.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 2.1 | 55.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 2.1 | 29.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 2.1 | 26.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 1.9 | 26.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 1.8 | 27.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 1.6 | 46.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 1.6 | 44.2 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 1.6 | 85.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 1.6 | 38.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 1.5 | 32.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 1.4 | 25.8 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 1.4 | 17.7 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 1.3 | 21.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 1.1 | 18.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 1.1 | 14.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.9 | 149.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.9 | 37.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.9 | 16.2 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.8 | 74.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.8 | 69.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.7 | 13.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.6 | 6.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.6 | 36.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.5 | 17.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.5 | 7.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.4 | 43.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.4 | 4.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.4 | 29.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.4 | 30.0 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.3 | 5.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 6.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 6.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 4.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 8.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.2 | 9.3 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.1 | 5.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 8.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 9.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 5.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |