Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
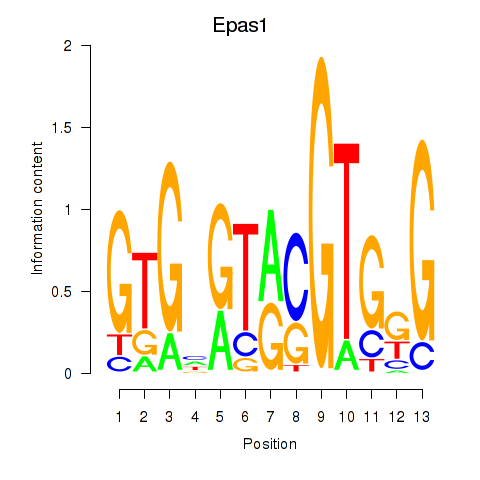
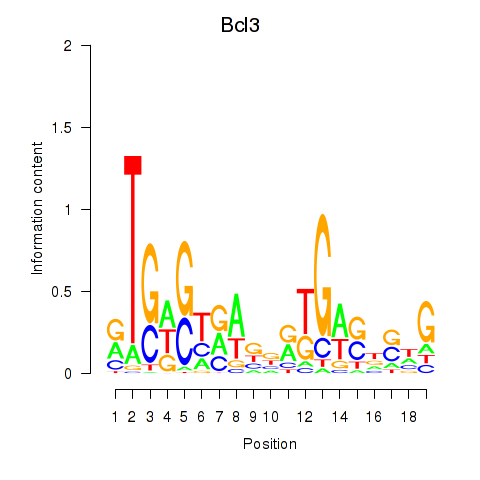
Results for Epas1_Bcl3
Z-value: 1.98
Transcription factors associated with Epas1_Bcl3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Epas1
|
ENSRNOG00000021318 | endothelial PAS domain protein 1 |
|
Bcl3
|
ENSRNOG00000043416 | B-cell CLL/lymphoma 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl3 | rn6_v1_chr1_-_80744831_80744831 | -0.44 | 1.6e-16 | Click! |
| Epas1 | rn6_v1_chr6_+_10306405_10306508 | 0.28 | 2.4e-07 | Click! |
Activity profile of Epas1_Bcl3 motif
Sorted Z-values of Epas1_Bcl3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_18079560 | 165.73 |
ENSRNOT00000072093
|
AABR07031533.1
|
|
| chr8_+_39305128 | 154.75 |
ENSRNOT00000008285
|
Fez1
|
fasciculation and elongation protein zeta 1 |
| chr5_-_130085838 | 119.52 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr7_+_45328105 | 104.58 |
ENSRNOT00000029208
ENSRNOT00000090641 |
Slc6a15
|
solute carrier family 6 member 15 |
| chr2_-_179704629 | 100.67 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr10_+_80790168 | 97.61 |
ENSRNOT00000073315
ENSRNOT00000075163 |
Car10
|
carbonic anhydrase 10 |
| chr2_+_18354542 | 90.03 |
ENSRNOT00000042958
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr9_+_73334618 | 89.05 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr3_+_71210301 | 88.11 |
ENSRNOT00000006504
|
Fam171b
|
family with sequence similarity 171, member B |
| chr2_-_257864385 | 82.89 |
ENSRNOT00000072048
|
Ak5
|
adenylate kinase 5 |
| chr1_+_226435979 | 82.17 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr3_-_64554953 | 76.48 |
ENSRNOT00000067452
|
LOC102553814
|
serine/arginine repetitive matrix protein 1-like |
| chr8_-_94563760 | 75.41 |
ENSRNOT00000032792
|
Snap91
|
synaptosomal-associated protein 91 |
| chrX_+_151103576 | 73.08 |
ENSRNOT00000015401
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr15_-_52317219 | 62.56 |
ENSRNOT00000016555
|
Dmtn
|
dematin actin binding protein |
| chr14_+_99529284 | 59.07 |
ENSRNOT00000006897
ENSRNOT00000067134 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr10_-_8498422 | 56.39 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr4_+_21317695 | 56.18 |
ENSRNOT00000007572
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr12_+_31934343 | 54.48 |
ENSRNOT00000011181
|
Tmem132d
|
transmembrane protein 132D |
| chr3_+_97723901 | 53.05 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr1_+_192613372 | 50.69 |
ENSRNOT00000016632
|
Cacng3
|
calcium voltage-gated channel auxiliary subunit gamma 3 |
| chr6_+_83083740 | 49.28 |
ENSRNOT00000007600
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr15_-_14737704 | 48.47 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
| chr10_+_27973681 | 47.67 |
ENSRNOT00000004970
|
Gabrb2
|
gamma-aminobutyric acid type A receptor beta 2 subunit |
| chrX_-_124464963 | 47.24 |
ENSRNOT00000036472
ENSRNOT00000077697 |
Tmem255a
|
transmembrane protein 255A |
| chr7_-_93280009 | 47.22 |
ENSRNOT00000064877
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr17_-_6244612 | 46.84 |
ENSRNOT00000042145
ENSRNOT00000090914 ENSRNOT00000082611 |
Ntrk2
|
neurotrophic receptor tyrosine kinase 2 |
| chr3_-_52510507 | 46.79 |
ENSRNOT00000091259
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr1_-_260992291 | 46.17 |
ENSRNOT00000035415
ENSRNOT00000034758 |
Slit1
|
slit guidance ligand 1 |
| chr4_+_161720501 | 44.49 |
ENSRNOT00000008196
ENSRNOT00000055886 |
Nrip2
|
nuclear receptor interacting protein 2 |
| chr5_+_148661070 | 43.61 |
ENSRNOT00000056229
|
Nkain1
|
Sodium/potassium transporting ATPase interacting 1 |
| chr4_-_144869919 | 43.37 |
ENSRNOT00000009066
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chrX_-_142131545 | 42.88 |
ENSRNOT00000077402
|
Fgf13
|
fibroblast growth factor 13 |
| chr7_+_79638046 | 42.78 |
ENSRNOT00000029419
|
Zfpm2
|
zinc finger protein, multitype 2 |
| chr15_+_34187223 | 42.08 |
ENSRNOT00000024978
|
Cpne6
|
copine 6 |
| chr10_-_85084850 | 42.02 |
ENSRNOT00000012462
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr5_+_31568419 | 41.17 |
ENSRNOT00000050137
|
Mmp16
|
matrix metallopeptidase 16 |
| chr10_+_17261541 | 40.96 |
ENSRNOT00000005478
|
Sh3pxd2b
|
SH3 and PX domains 2B |
| chr8_-_110813000 | 40.62 |
ENSRNOT00000010634
|
Ephb1
|
Eph receptor B1 |
| chr4_+_29978739 | 39.67 |
ENSRNOT00000011756
|
Ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr9_+_73529612 | 38.56 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr18_+_56193978 | 38.09 |
ENSRNOT00000041533
ENSRNOT00000080177 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr1_+_82174451 | 38.01 |
ENSRNOT00000027783
|
Tmem145
|
transmembrane protein 145 |
| chr18_+_59748444 | 37.58 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr8_+_27777179 | 37.29 |
ENSRNOT00000009825
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 |
| chr1_-_212548730 | 35.66 |
ENSRNOT00000089729
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr1_+_72860218 | 35.64 |
ENSRNOT00000024547
|
Syt5
|
synaptotagmin 5 |
| chr15_-_34550850 | 35.05 |
ENSRNOT00000027794
ENSRNOT00000090228 |
Cbln3
|
cerebellin 3 precursor |
| chr5_-_7941822 | 34.64 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr7_+_123168811 | 34.61 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr20_+_3558827 | 34.26 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr8_+_97277456 | 32.98 |
ENSRNOT00000082757
|
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr4_-_114853868 | 32.74 |
ENSRNOT00000090828
|
Wdr54
|
WD repeat domain 54 |
| chr3_+_177225737 | 32.49 |
ENSRNOT00000045845
|
Oprl1
|
opioid related nociceptin receptor 1 |
| chr1_+_170262156 | 32.18 |
ENSRNOT00000024077
ENSRNOT00000085395 |
Cckbr
|
cholecystokinin B receptor |
| chr5_+_136683592 | 31.51 |
ENSRNOT00000085527
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr4_+_41364441 | 31.00 |
ENSRNOT00000087146
|
Foxp2
|
forkhead box P2 |
| chr7_-_112833083 | 30.32 |
ENSRNOT00000006966
|
Fam135b
|
family with sequence similarity 135, member B |
| chr15_-_58711872 | 30.26 |
ENSRNOT00000058204
|
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr10_-_89700283 | 30.17 |
ENSRNOT00000028213
|
Etv4
|
ets variant 4 |
| chr5_-_62621737 | 30.00 |
ENSRNOT00000011573
|
Gabbr2
|
gamma-aminobutyric acid type B receptor subunit 2 |
| chr2_+_198721724 | 29.71 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr2_-_96668222 | 28.63 |
ENSRNOT00000016567
|
Pkia
|
cAMP-dependent protein kinase inhibitor alpha |
| chr6_+_126434226 | 28.31 |
ENSRNOT00000090857
|
Chga
|
chromogranin A |
| chr3_-_82856171 | 28.18 |
ENSRNOT00000088555
|
RGD1564664
|
similar to LOC387763 protein |
| chr6_-_39363367 | 28.18 |
ENSRNOT00000088687
ENSRNOT00000065531 |
Fam84a
|
family with sequence similarity 84, member A |
| chr19_+_50848736 | 28.09 |
ENSRNOT00000077053
|
Cdh13
|
cadherin 13 |
| chr16_+_23668595 | 27.39 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_+_29191192 | 27.30 |
ENSRNOT00000018718
|
Hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr4_-_160334910 | 27.06 |
ENSRNOT00000086772
|
Prmt8
|
protein arginine methyltransferase 8 |
| chr5_-_145418992 | 26.80 |
ENSRNOT00000037128
|
Gjb4
|
gap junction protein, beta 4 |
| chr8_+_408001 | 26.52 |
ENSRNOT00000046058
|
Gucy1a2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr5_-_62186372 | 26.08 |
ENSRNOT00000089789
|
Coro2a
|
coronin 2A |
| chr13_-_105141030 | 26.06 |
ENSRNOT00000003313
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr1_+_89491654 | 25.96 |
ENSRNOT00000028632
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr1_+_80954858 | 25.59 |
ENSRNOT00000030440
|
Zfp112
|
zinc finger protein 112 |
| chr5_-_34813116 | 25.55 |
ENSRNOT00000017479
|
Nkain3
|
Sodium/potassium transporting ATPase interacting 3 |
| chr6_-_136145837 | 25.29 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
| chr1_-_59732409 | 25.18 |
ENSRNOT00000014824
|
Has1
|
hyaluronan synthase 1 |
| chr1_+_87248489 | 25.01 |
ENSRNOT00000028091
|
Dpf1
|
double PHD fingers 1 |
| chr1_-_72727112 | 24.92 |
ENSRNOT00000031172
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr5_+_60809762 | 24.87 |
ENSRNOT00000016839
|
Frmpd1
|
FERM and PDZ domain containing 1 |
| chr13_-_49313940 | 24.36 |
ENSRNOT00000012190
|
Cntn2
|
contactin 2 |
| chr3_+_171832500 | 24.23 |
ENSRNOT00000007554
|
Vapb
|
VAMP associated protein B and C |
| chr4_+_22445414 | 24.04 |
ENSRNOT00000087657
ENSRNOT00000030224 |
Rundc3b
|
RUN domain containing 3B |
| chr11_-_84008946 | 24.00 |
ENSRNOT00000037612
|
Vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr14_+_36071376 | 23.97 |
ENSRNOT00000082183
|
Lnx1
|
ligand of numb-protein X 1 |
| chr10_-_90994437 | 23.71 |
ENSRNOT00000093167
|
Gfap
|
glial fibrillary acidic protein |
| chr11_+_60383431 | 23.66 |
ENSRNOT00000093295
|
Cd200
|
Cd200 molecule |
| chr8_-_84522588 | 23.35 |
ENSRNOT00000076213
|
Mlip
|
muscular LMNA-interacting protein |
| chr4_-_125958112 | 22.71 |
ENSRNOT00000071315
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr2_-_181026024 | 22.34 |
ENSRNOT00000064930
|
Gucy1b3
|
guanylate cyclase 1 soluble subunit beta 3 |
| chr2_+_236625357 | 21.97 |
ENSRNOT00000081248
|
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr18_-_37819097 | 21.93 |
ENSRNOT00000025880
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr1_+_175586097 | 21.36 |
ENSRNOT00000024933
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr17_-_9792007 | 21.35 |
ENSRNOT00000021596
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr14_+_17534412 | 20.69 |
ENSRNOT00000079304
ENSRNOT00000046771 |
Cdkl2
|
cyclin dependent kinase like 2 |
| chr2_-_173563273 | 20.54 |
ENSRNOT00000081423
|
Zbbx
|
zinc finger, B-box domain containing |
| chr1_-_220729000 | 20.48 |
ENSRNOT00000027727
|
Cst6
|
cystatin E/M |
| chr3_+_160207913 | 20.39 |
ENSRNOT00000014346
|
Wisp2
|
WNT1 inducible signaling pathway protein 2 |
| chr5_-_58078545 | 20.33 |
ENSRNOT00000075777
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr5_-_160179978 | 20.08 |
ENSRNOT00000022820
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr7_+_141326321 | 20.02 |
ENSRNOT00000088191
|
Asic1
|
acid sensing ion channel subunit 1 |
| chr18_+_17043903 | 20.00 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr20_-_31956649 | 19.95 |
ENSRNOT00000072429
|
Hk1
|
hexokinase 1 |
| chr3_-_54593306 | 19.85 |
ENSRNOT00000087122
|
Stk39
|
serine threonine kinase 39 |
| chr15_-_100348760 | 19.80 |
ENSRNOT00000085607
|
AABR07019341.1
|
|
| chr4_-_45332420 | 19.55 |
ENSRNOT00000083039
|
Wnt2
|
wingless-type MMTV integration site family member 2 |
| chr7_-_76035096 | 19.23 |
ENSRNOT00000072255
|
AABR07057530.1
|
|
| chr2_+_58448917 | 18.42 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr1_-_89488223 | 17.89 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr5_-_7874909 | 17.82 |
ENSRNOT00000064774
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_+_202432366 | 17.63 |
ENSRNOT00000027681
|
Plpp4
|
phospholipid phosphatase 4 |
| chr15_-_13228607 | 17.57 |
ENSRNOT00000042010
ENSRNOT00000088214 |
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chr10_-_44746549 | 17.46 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chr15_+_108908607 | 17.39 |
ENSRNOT00000089455
|
Zic2
|
Zic family member 2 |
| chr9_+_82700468 | 17.03 |
ENSRNOT00000027227
|
Inha
|
inhibin alpha subunit |
| chr1_+_251633199 | 16.87 |
ENSRNOT00000084785
|
AC094647.2
|
|
| chr20_-_6500523 | 16.53 |
ENSRNOT00000000629
|
Cpne5
|
copine 5 |
| chr19_-_46101250 | 16.48 |
ENSRNOT00000015874
|
Adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr7_+_83113672 | 16.18 |
ENSRNOT00000006783
|
Trhr
|
thyrotropin releasing hormone receptor |
| chr4_-_117153907 | 16.14 |
ENSRNOT00000091374
|
Rab11fip5
|
RAB11 family interacting protein 5 |
| chr7_-_118396728 | 16.03 |
ENSRNOT00000066431
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr4_-_157372861 | 15.96 |
ENSRNOT00000021578
|
P3h3
|
prolyl 3-hydroxylase 3 |
| chr3_+_110442637 | 15.85 |
ENSRNOT00000010471
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr10_+_90731148 | 15.67 |
ENSRNOT00000093604
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr6_+_102311116 | 15.67 |
ENSRNOT00000083433
|
Arg2
|
arginase 2 |
| chr5_-_68139199 | 15.66 |
ENSRNOT00000009987
|
Plppr1
|
phospholipid phosphatase related 1 |
| chr10_+_10555337 | 15.40 |
ENSRNOT00000032495
|
RGD1565166
|
similar to MGC45438 protein |
| chr2_+_239415046 | 15.25 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr1_+_84328114 | 14.96 |
ENSRNOT00000028266
|
Hipk4
|
homeodomain interacting protein kinase 4 |
| chr14_+_17064353 | 14.90 |
ENSRNOT00000003052
|
Scarb2
|
scavenger receptor class B, member 2 |
| chr12_-_5682608 | 14.38 |
ENSRNOT00000076483
|
Fry
|
FRY microtubule binding protein |
| chr1_+_80953275 | 14.07 |
ENSRNOT00000087253
|
Zfp112
|
zinc finger protein 112 |
| chr2_-_210726179 | 14.05 |
ENSRNOT00000025689
|
Gstm7
|
glutathione S-transferase, mu 7 |
| chr15_+_52241801 | 13.83 |
ENSRNOT00000082639
|
Hr
|
HR, lysine demethylase and nuclear receptor corepressor |
| chr3_+_3767394 | 13.62 |
ENSRNOT00000067840
|
Gpsm1
|
G-protein signaling modulator 1 |
| chr13_-_92988137 | 13.35 |
ENSRNOT00000004803
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr14_-_20920286 | 13.17 |
ENSRNOT00000004391
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr14_-_21127868 | 13.05 |
ENSRNOT00000020608
ENSRNOT00000092172 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr11_+_80736576 | 12.49 |
ENSRNOT00000047678
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr10_-_105628091 | 12.46 |
ENSRNOT00000016067
|
Cygb
|
cytoglobin |
| chr7_-_123445613 | 12.33 |
ENSRNOT00000070937
|
Shisa8
|
shisa family member 8 |
| chr10_-_89699836 | 11.99 |
ENSRNOT00000084311
|
Etv4
|
ets variant 4 |
| chr10_+_46783979 | 11.79 |
ENSRNOT00000005237
|
Gid4
|
GID complex subunit 4 |
| chr1_+_192379543 | 11.71 |
ENSRNOT00000078705
|
Prkcb
|
protein kinase C, beta |
| chr8_-_7426611 | 11.68 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr12_+_48598647 | 11.49 |
ENSRNOT00000000889
|
Tmem119
|
transmembrane protein 119 |
| chr15_-_42898150 | 11.40 |
ENSRNOT00000030036
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr2_-_5579894 | 11.17 |
ENSRNOT00000020044
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr10_-_88389347 | 11.09 |
ENSRNOT00000022695
|
Klhl11
|
kelch-like family member 11 |
| chr15_+_102164751 | 10.95 |
ENSRNOT00000076400
|
Gpc6
|
glypican 6 |
| chr1_+_140792049 | 10.87 |
ENSRNOT00000049681
|
Acan
|
aggrecan |
| chr1_+_219250265 | 10.73 |
ENSRNOT00000024353
|
Doc2g
|
double C2-like domains, gamma |
| chr1_+_80920747 | 10.48 |
ENSRNOT00000026077
|
Zfp180
|
zinc finger protein 180 |
| chr9_-_44419998 | 10.39 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr16_-_930527 | 10.34 |
ENSRNOT00000014787
|
Spin1
|
spindlin 1 |
| chr3_+_111160205 | 10.18 |
ENSRNOT00000019392
|
Chac1
|
ChaC glutathione-specific gamma-glutamylcyclotransferase 1 |
| chr1_-_266333105 | 9.92 |
ENSRNOT00000027093
|
Arl3
|
ADP ribosylation factor like GTPase 3 |
| chr7_+_71023976 | 9.77 |
ENSRNOT00000005679
|
Tac3
|
tachykinin 3 |
| chr8_-_45137893 | 9.71 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr3_-_162872831 | 9.53 |
ENSRNOT00000008478
|
Sulf2
|
sulfatase 2 |
| chrX_-_70978952 | 9.44 |
ENSRNOT00000076910
|
Slc7a3
|
solute carrier family 7 member 3 |
| chr1_+_165625058 | 9.39 |
ENSRNOT00000025104
|
Rab6a
|
RAB6A, member RAS oncogene family |
| chr5_+_16463597 | 9.25 |
ENSRNOT00000091550
|
AABR07047036.1
|
|
| chr14_+_104250617 | 9.25 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr1_+_165625350 | 9.24 |
ENSRNOT00000087534
|
Rab6a
|
RAB6A, member RAS oncogene family |
| chr8_-_123829749 | 9.24 |
ENSRNOT00000090467
|
AABR07071598.1
|
|
| chr1_+_164502389 | 8.52 |
ENSRNOT00000043554
|
Arrb1
|
arrestin, beta 1 |
| chr20_+_6102277 | 8.50 |
ENSRNOT00000033064
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chrX_-_123601100 | 8.33 |
ENSRNOT00000092546
ENSRNOT00000092301 |
Sept6
|
septin 6 |
| chr3_-_12029877 | 8.26 |
ENSRNOT00000022327
|
Slc2a8
|
solute carrier family 2 member 8 |
| chr5_+_148923098 | 8.07 |
ENSRNOT00000048781
|
Sdc3
|
syndecan 3 |
| chr6_+_21708487 | 7.80 |
ENSRNOT00000087358
|
AABR07063197.1
|
|
| chr2_+_30094421 | 7.61 |
ENSRNOT00000082226
|
AABR07007839.1
|
|
| chr2_+_2694480 | 7.51 |
ENSRNOT00000083565
|
AABR07072671.1
|
|
| chr1_-_188895223 | 7.48 |
ENSRNOT00000032796
|
Gpr139
|
G protein-coupled receptor 139 |
| chr16_-_755990 | 7.47 |
ENSRNOT00000013501
|
Polr3a
|
RNA polymerase III subunit A |
| chr9_-_20528879 | 7.37 |
ENSRNOT00000085293
|
AABR07066871.3
|
|
| chr17_-_9791781 | 7.32 |
ENSRNOT00000090536
|
Rgs14
|
regulator of G-protein signaling 14 |
| chrX_+_135348436 | 6.95 |
ENSRNOT00000008868
|
Rab33a
|
RAB33A, member RAS oncogene family |
| chr6_-_111015598 | 6.91 |
ENSRNOT00000015046
|
Zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr3_-_46153371 | 6.89 |
ENSRNOT00000085885
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr17_-_77687456 | 6.89 |
ENSRNOT00000045765
ENSRNOT00000081645 |
Frmd4a
|
FERM domain containing 4A |
| chr1_-_154111725 | 6.80 |
ENSRNOT00000055488
|
Ccdc81
|
coiled-coil domain containing 81 |
| chr7_+_25808419 | 6.75 |
ENSRNOT00000091571
|
Rfx4
|
regulatory factor X4 |
| chr3_+_138770574 | 6.51 |
ENSRNOT00000012510
ENSRNOT00000066986 |
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr1_+_100577056 | 6.49 |
ENSRNOT00000026992
|
Napsa
|
napsin A aspartic peptidase |
| chr16_-_81583916 | 6.45 |
ENSRNOT00000026281
|
LOC103693999
|
transmembrane and coiled-coil domain-containing protein 3 |
| chr17_-_1085885 | 6.24 |
ENSRNOT00000026287
|
Ptch1
|
patched 1 |
| chr7_+_97559841 | 6.18 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr20_+_47779056 | 6.09 |
ENSRNOT00000090015
|
AC132752.1
|
|
| chr10_-_74070266 | 6.08 |
ENSRNOT00000005987
|
Cltc
|
clathrin heavy chain |
| chr10_+_14136883 | 5.82 |
ENSRNOT00000082295
|
AC115181.1
|
|
| chr10_+_37724915 | 5.75 |
ENSRNOT00000008477
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr2_+_93827504 | 5.60 |
ENSRNOT00000032059
|
Pmp2
|
peripheral myelin protein 2 |
| chr6_+_135610743 | 5.59 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr17_+_21490402 | 5.57 |
ENSRNOT00000020244
|
Gcm2
|
glial cells missing homolog 2 |
| chr16_-_6675746 | 5.47 |
ENSRNOT00000025858
|
Prkcd
|
protein kinase C, delta |
| chr15_+_103344476 | 5.20 |
ENSRNOT00000013329
ENSRNOT00000077047 ENSRNOT00000078103 |
Gpr180
|
G protein-coupled receptor 180 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Epas1_Bcl3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 34.9 | 104.6 | GO:0015820 | leucine transport(GO:0015820) |
| 27.4 | 82.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 15.6 | 62.6 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 14.3 | 42.8 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 13.3 | 53.4 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 12.9 | 154.7 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 12.5 | 37.6 | GO:1990743 | protein sialylation(GO:1990743) |
| 11.8 | 59.1 | GO:0070343 | white fat cell proliferation(GO:0070343) positive regulation of fat cell proliferation(GO:0070346) regulation of white fat cell proliferation(GO:0070350) |
| 11.5 | 46.2 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 10.8 | 75.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 9.6 | 28.7 | GO:0010070 | zygote asymmetric cell division(GO:0010070) |
| 9.4 | 28.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 8.1 | 32.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 8.1 | 24.4 | GO:0060167 | clustering of voltage-gated potassium channels(GO:0045163) regulation of adenosine receptor signaling pathway(GO:0060167) |
| 7.6 | 38.1 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) NMDA selective glutamate receptor signaling pathway(GO:0098989) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 6.8 | 41.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) positive regulation of adipose tissue development(GO:1904179) |
| 6.7 | 46.8 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 6.6 | 19.9 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 6.6 | 46.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 6.6 | 39.7 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 6.5 | 19.5 | GO:1904953 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 6.5 | 84.1 | GO:1990834 | response to odorant(GO:1990834) |
| 6.3 | 25.2 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 5.1 | 56.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 5.0 | 19.9 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 4.8 | 24.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 4.7 | 18.6 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 4.6 | 100.7 | GO:0060992 | response to fungicide(GO:0060992) |
| 4.5 | 27.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 4.5 | 17.9 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 4.5 | 13.4 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 4.4 | 31.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 4.3 | 17.4 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 4.1 | 20.3 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 4.0 | 23.7 | GO:0010625 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 3.9 | 11.7 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 3.9 | 11.7 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 3.8 | 11.4 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 3.6 | 46.8 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 3.6 | 14.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 3.5 | 17.6 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 3.4 | 47.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 3.4 | 40.6 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 3.2 | 9.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 3.1 | 22.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 3.1 | 15.7 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 3.1 | 34.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 3.0 | 6.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 3.0 | 23.7 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 2.9 | 31.5 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 2.9 | 20.0 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 2.8 | 25.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 2.8 | 22.3 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 2.7 | 16.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 2.7 | 26.8 | GO:0042048 | olfactory behavior(GO:0042048) |
| 2.6 | 28.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 2.4 | 17.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 2.4 | 89.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 2.3 | 56.4 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 2.3 | 83.7 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 2.3 | 32.2 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 2.3 | 16.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 2.2 | 35.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 2.1 | 17.2 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 2.1 | 8.5 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 2.1 | 6.2 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 2.0 | 20.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 1.9 | 9.4 | GO:1902022 | lysine transport(GO:0015819) L-lysine transport(GO:1902022) L-lysine transmembrane transport(GO:1903401) |
| 1.9 | 37.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 1.8 | 5.5 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 1.7 | 26.5 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 1.6 | 11.5 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 1.6 | 9.8 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 1.6 | 50.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 1.6 | 16.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 1.6 | 119.5 | GO:0042220 | response to cocaine(GO:0042220) |
| 1.6 | 12.5 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 1.5 | 13.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 1.5 | 21.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 1.4 | 6.8 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 1.3 | 46.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 1.3 | 10.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 1.2 | 3.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 1.2 | 4.9 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 1.2 | 4.7 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 1.2 | 30.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 1.1 | 27.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 1.1 | 16.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 1.0 | 12.5 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 1.0 | 52.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 1.0 | 73.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 1.0 | 23.4 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.9 | 9.5 | GO:0009169 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.9 | 11.4 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.9 | 11.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.8 | 33.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.8 | 3.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.8 | 5.6 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.8 | 4.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.8 | 5.6 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.8 | 23.0 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.8 | 1.6 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.8 | 12.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.7 | 16.7 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.7 | 2.8 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.7 | 13.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.7 | 59.0 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.7 | 54.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.7 | 9.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.7 | 13.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.7 | 1.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.6 | 3.2 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.6 | 5.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.6 | 2.2 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.5 | 2.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.5 | 1.6 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.5 | 33.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.5 | 42.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.5 | 15.0 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.5 | 3.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.4 | 5.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.4 | 2.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.4 | 13.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.4 | 2.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.3 | 4.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.3 | 5.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.3 | 28.6 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.3 | 2.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 6.9 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.3 | 2.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.3 | 6.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.3 | 10.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.3 | 28.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.3 | 9.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.3 | 44.5 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.3 | 1.4 | GO:0006566 | L-serine biosynthetic process(GO:0006564) threonine metabolic process(GO:0006566) |
| 0.3 | 2.9 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.3 | 8.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 6.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 2.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.2 | 2.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.2 | 4.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 3.4 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 2.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 4.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 | 15.9 | GO:0007613 | memory(GO:0007613) |
| 0.1 | 1.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 16.4 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 9.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 20.1 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.1 | 1.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 7.5 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 3.4 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.1 | 25.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.4 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 5.0 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.8 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 25.1 | 75.4 | GO:0098830 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 20.9 | 62.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 12.0 | 119.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 10.0 | 30.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 9.9 | 39.7 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 9.9 | 89.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 9.5 | 38.1 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 8.4 | 100.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 7.5 | 82.2 | GO:0032009 | early phagosome(GO:0032009) |
| 7.1 | 35.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 6.8 | 20.3 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 5.9 | 23.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 5.7 | 17.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 3.3 | 56.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 3.1 | 37.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 2.7 | 71.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 2.6 | 28.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 2.3 | 47.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 2.2 | 48.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 2.2 | 10.9 | GO:0072534 | perineuronal net(GO:0072534) |
| 2.1 | 40.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 2.1 | 63.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 2.0 | 48.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.8 | 27.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 1.8 | 44.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 1.7 | 8.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.7 | 25.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 1.5 | 4.5 | GO:0032173 | septin collar(GO:0032173) |
| 1.5 | 48.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 1.4 | 16.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 1.3 | 26.8 | GO:0005922 | connexon complex(GO:0005922) |
| 1.2 | 24.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.9 | 46.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.8 | 20.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.7 | 42.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.7 | 18.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.7 | 8.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.7 | 8.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.6 | 3.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.6 | 30.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.6 | 6.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.6 | 19.9 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.6 | 26.8 | GO:0002102 | podosome(GO:0002102) |
| 0.5 | 193.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.5 | 191.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.5 | 5.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.5 | 6.5 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.4 | 76.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.4 | 7.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.4 | 35.1 | GO:0005901 | caveola(GO:0005901) |
| 0.3 | 4.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 34.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.3 | 2.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 49.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 4.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 0.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 3.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 25.4 | GO:0005903 | brush border(GO:0005903) |
| 0.2 | 4.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 9.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 85.7 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.1 | 23.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 8.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 5.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 8.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 5.8 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 2.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 2.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 3.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 4.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 3.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 9.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 4.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.1 | 100.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 18.7 | 56.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 12.5 | 37.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 12.4 | 37.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 10.5 | 104.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 9.4 | 46.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 9.1 | 27.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 8.4 | 83.9 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 8.3 | 41.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 7.5 | 82.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 7.5 | 30.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 7.3 | 22.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 7.0 | 28.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 6.8 | 40.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 6.7 | 20.0 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 6.6 | 46.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 6.1 | 24.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 5.4 | 32.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 5.4 | 27.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 5.3 | 16.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 5.3 | 185.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 5.2 | 26.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 5.1 | 20.3 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 5.0 | 25.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 4.9 | 34.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 4.4 | 119.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 4.2 | 12.5 | GO:0004096 | catalase activity(GO:0004096) |
| 4.0 | 24.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 3.9 | 11.7 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 3.9 | 89.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 3.6 | 25.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 3.6 | 100.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 3.5 | 31.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 3.5 | 38.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 3.4 | 47.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 3.3 | 19.9 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 2.9 | 41.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 2.9 | 159.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 2.8 | 42.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 2.5 | 10.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 2.4 | 28.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 2.4 | 52.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 2.4 | 9.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 2.1 | 6.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 2.0 | 51.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 2.0 | 33.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 2.0 | 48.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 1.8 | 5.5 | GO:0070976 | TIR domain binding(GO:0070976) |
| 1.8 | 41.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 1.7 | 46.8 | GO:0031402 | sodium ion binding(GO:0031402) |
| 1.6 | 11.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 1.6 | 9.4 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 1.5 | 39.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 1.4 | 11.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.4 | 4.1 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 1.4 | 2.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 1.3 | 9.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.3 | 63.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 1.3 | 13.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 1.2 | 8.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 1.2 | 4.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.2 | 15.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.1 | 53.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.1 | 27.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 1.0 | 8.1 | GO:0036122 | BMP binding(GO:0036122) |
| 1.0 | 19.5 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 1.0 | 50.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.9 | 31.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.8 | 18.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.8 | 32.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.8 | 35.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.8 | 20.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.7 | 8.3 | GO:0005536 | glucose binding(GO:0005536) |
| 0.7 | 13.2 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.6 | 11.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.6 | 19.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.5 | 1.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.5 | 4.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 34.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.5 | 43.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.5 | 33.0 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.5 | 7.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.5 | 2.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.5 | 2.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.4 | 2.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 8.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 6.5 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.3 | 23.4 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.3 | 4.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 8.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 49.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.3 | 2.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 3.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.3 | 1.6 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 5.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 22.1 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.2 | 3.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 28.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 2.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 10.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 25.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.2 | 3.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 41.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.2 | 6.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 4.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 17.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 8.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.2 | 17.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.2 | 5.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 3.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 4.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 2.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 5.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 7.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 9.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 3.3 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.8 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 4.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 1.4 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 4.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 100.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 2.0 | 97.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 1.9 | 132.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 1.7 | 85.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 1.6 | 38.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.4 | 83.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 1.4 | 25.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.4 | 40.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 1.2 | 11.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.8 | 23.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.8 | 8.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.6 | 11.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 15.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.5 | 15.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.4 | 14.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.4 | 19.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.3 | 6.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.3 | 4.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 5.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 2.7 | ST ADRENERGIC | Adrenergic Pathway |
| 0.2 | 8.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 1.6 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.2 | 10.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 32.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 139.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 5.5 | 33.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 4.1 | 82.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 3.9 | 86.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 2.8 | 47.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 2.4 | 62.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 2.1 | 56.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 1.8 | 30.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 1.7 | 22.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.6 | 48.9 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 1.5 | 26.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 1.3 | 25.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 1.2 | 4.9 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 1.2 | 21.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.2 | 23.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 1.2 | 59.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 1.1 | 12.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.1 | 46.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 1.1 | 24.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 1.0 | 18.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 1.0 | 6.1 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.9 | 25.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.9 | 10.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.9 | 11.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.9 | 8.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.8 | 22.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.7 | 17.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 24.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.5 | 13.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.4 | 8.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 15.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 19.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.4 | 5.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.4 | 35.6 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.4 | 7.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.4 | 5.5 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.3 | 23.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.3 | 4.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 9.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 3.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 42.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 4.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 20.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 29.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 13.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 18.4 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.2 | 3.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 5.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 8.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.2 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 2.2 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |