Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
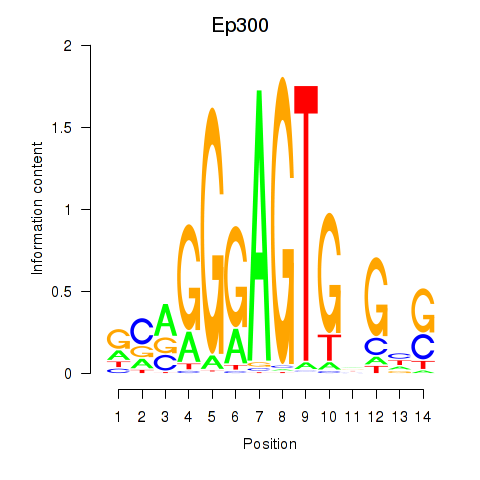
Results for Ep300
Z-value: 0.58
Transcription factors associated with Ep300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ep300
|
ENSRNOG00000000190 | E1A binding protein p300 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ep300 | rn6_v1_chr7_+_122818975_122818975 | 0.49 | 4.1e-21 | Click! |
Activity profile of Ep300 motif
Sorted Z-values of Ep300 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_1339488 | 15.47 |
ENSRNOT00000041074
|
RT1-M2
|
RT1 class Ib, locus M2 |
| chr1_-_199823386 | 13.50 |
ENSRNOT00000027375
|
Rgs10
|
regulator of G-protein signaling 10 |
| chr14_+_91783514 | 12.35 |
ENSRNOT00000080753
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr16_-_9430743 | 11.09 |
ENSRNOT00000043811
|
Wdfy4
|
WDFY family member 4 |
| chr4_-_119327822 | 9.88 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr12_+_36694960 | 9.54 |
ENSRNOT00000064276
ENSRNOT00000001299 |
Scarb1
|
scavenger receptor class B, member 1 |
| chr6_-_103470427 | 9.49 |
ENSRNOT00000091560
ENSRNOT00000088795 ENSRNOT00000079824 |
Actn1
|
actinin, alpha 1 |
| chr7_-_12634641 | 8.59 |
ENSRNOT00000093132
|
Cfd
|
complement factor D |
| chr9_-_16795887 | 8.43 |
ENSRNOT00000071609
|
Cd79al
|
Cd79a molecule-like |
| chrX_+_15273933 | 7.36 |
ENSRNOT00000075082
|
LOC108348091
|
erythroid transcription factor |
| chr1_-_167347490 | 6.92 |
ENSRNOT00000076499
|
Rhog
|
ras homolog family member G |
| chr20_+_3230052 | 6.62 |
ENSRNOT00000078454
|
RT1-T24-3
|
RT1 class I, locus T24, gene 3 |
| chr20_-_22004209 | 6.59 |
ENSRNOT00000086250
ENSRNOT00000068778 |
Rtkn2
|
rhotekin 2 |
| chr20_-_1980101 | 6.58 |
ENSRNOT00000084582
ENSRNOT00000085050 ENSRNOT00000082545 ENSRNOT00000088396 |
Gabbr1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr10_+_64360390 | 6.15 |
ENSRNOT00000010168
|
Fam57a
|
family with sequence similarity 57, member A |
| chr13_+_89774764 | 6.15 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr1_-_167347662 | 6.01 |
ENSRNOT00000027641
ENSRNOT00000076592 |
Rhog
|
ras homolog family member G |
| chr7_+_12471824 | 5.99 |
ENSRNOT00000068197
|
Sbno2
|
strawberry notch homolog 2 |
| chr8_-_21995806 | 5.84 |
ENSRNOT00000028034
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr7_-_59514939 | 5.79 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr14_+_22107416 | 5.69 |
ENSRNOT00000060179
ENSRNOT00000080114 |
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr4_-_170117325 | 5.42 |
ENSRNOT00000074198
|
LOC100912564
|
histone H4-like |
| chrX_+_156463953 | 5.24 |
ENSRNOT00000079889
|
Flna
|
filamin A |
| chr11_+_27364916 | 5.23 |
ENSRNOT00000002151
|
Bach1
|
BTB domain and CNC homolog 1 |
| chr14_-_34115273 | 5.18 |
ENSRNOT00000032156
|
Cep135
|
centrosomal protein 135 |
| chr3_+_159995064 | 5.18 |
ENSRNOT00000012606
|
Ttpal
|
alpha tocopherol transfer protein like |
| chr4_-_170145262 | 5.15 |
ENSRNOT00000074526
|
Hist1h4b
|
histone cluster 1, H4b |
| chr7_-_28040510 | 5.09 |
ENSRNOT00000005674
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr10_+_39109522 | 5.07 |
ENSRNOT00000010968
|
Irf1
|
interferon regulatory factor 1 |
| chr18_-_1389929 | 4.84 |
ENSRNOT00000051362
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr1_-_234670113 | 4.83 |
ENSRNOT00000017133
|
LOC499331
|
similar to hypothetical protein D030056L22 |
| chr1_+_173252058 | 4.78 |
ENSRNOT00000073421
|
LOC499229
|
similar to very large inducible GTPase 1 isoform A |
| chr10_-_102269272 | 4.61 |
ENSRNOT00000048264
|
Cdc42ep4
|
CDC42 effector protein 4 |
| chr15_-_51839341 | 4.50 |
ENSRNOT00000011277
|
RGD1308117
|
similar to 9930012K11Rik protein |
| chr6_-_107678156 | 4.45 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr8_+_44047592 | 4.36 |
ENSRNOT00000085084
|
Zfp202
|
zinc finger protein 202 |
| chr3_+_149221377 | 4.29 |
ENSRNOT00000016220
ENSRNOT00000087752 |
Mapre1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr20_+_30915213 | 4.29 |
ENSRNOT00000000681
|
Prf1
|
perforin 1 |
| chr11_+_50781127 | 4.18 |
ENSRNOT00000002738
|
Alcam
|
activated leukocyte cell adhesion molecule |
| chr4_-_115354795 | 3.98 |
ENSRNOT00000017691
|
Cd207
|
CD207 molecule |
| chr2_+_230901126 | 3.95 |
ENSRNOT00000016026
ENSRNOT00000015564 ENSRNOT00000068198 |
Camk2d
|
calcium/calmodulin-dependent protein kinase II delta |
| chr18_-_14016713 | 3.84 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr2_+_136993208 | 3.66 |
ENSRNOT00000040187
ENSRNOT00000066542 |
Pcdh10
|
protocadherin 10 |
| chr10_-_81666523 | 3.57 |
ENSRNOT00000003658
|
Nme1
|
NME/NM23 nucleoside diphosphate kinase 1 |
| chr5_+_48011864 | 3.51 |
ENSRNOT00000067610
|
Mdn1
|
midasin AAA ATPase 1 |
| chr1_+_198932870 | 3.33 |
ENSRNOT00000055003
|
Fbrs
|
fibrosin |
| chr9_+_82647071 | 3.22 |
ENSRNOT00000027135
|
Asic4
|
acid sensing ion channel subunit family member 4 |
| chr3_-_151258580 | 3.21 |
ENSRNOT00000026120
|
Edem2
|
ER degradation enhancing alpha-mannosidase like protein 2 |
| chr8_-_63034226 | 3.18 |
ENSRNOT00000043434
|
Pml
|
promyelocytic leukemia |
| chr2_+_112868707 | 3.16 |
ENSRNOT00000017805
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr5_-_137238354 | 3.15 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr8_-_21968415 | 3.15 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr18_-_1390190 | 3.13 |
ENSRNOT00000061777
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr19_+_54553419 | 3.09 |
ENSRNOT00000025392
|
Jph3
|
junctophilin 3 |
| chr5_+_48012231 | 2.95 |
ENSRNOT00000077040
|
Mdn1
|
midasin AAA ATPase 1 |
| chr6_+_91697109 | 2.77 |
ENSRNOT00000006355
|
Arf6
|
ADP-ribosylation factor 6 |
| chr10_-_65507581 | 2.75 |
ENSRNOT00000016759
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr11_-_70961358 | 2.74 |
ENSRNOT00000002427
|
Lmln
|
leishmanolysin like peptidase |
| chr1_-_13175876 | 2.68 |
ENSRNOT00000084870
|
Abracl
|
ABRA C-terminal like |
| chr1_-_261229046 | 2.58 |
ENSRNOT00000075531
|
Mms19
|
MMS19 homolog, cytosolic iron-sulfur assembly component |
| chr7_+_116997020 | 2.52 |
ENSRNOT00000078368
|
Zfp707l1
|
zinc finger protein 707-like 1 |
| chr10_-_62648844 | 2.31 |
ENSRNOT00000034282
|
Abhd15
|
abhydrolase domain containing 15 |
| chr18_-_56115593 | 2.21 |
ENSRNOT00000045041
|
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr20_+_26755911 | 2.21 |
ENSRNOT00000083017
|
Herc4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr2_+_189922996 | 2.21 |
ENSRNOT00000016499
|
S100a16
|
S100 calcium binding protein A16 |
| chr5_+_133865331 | 2.18 |
ENSRNOT00000035409
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr4_+_6559545 | 2.08 |
ENSRNOT00000067183
|
Prkag2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr10_-_57275708 | 1.94 |
ENSRNOT00000005370
|
Pfn1
|
profilin 1 |
| chr5_+_61474000 | 1.90 |
ENSRNOT00000013930
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr10_+_65507364 | 1.83 |
ENSRNOT00000016297
|
Sdf2
|
stromal cell derived factor 2 |
| chr7_+_117026097 | 1.82 |
ENSRNOT00000085758
|
Zfp707
|
zinc finger protein 707 |
| chrX_+_140878216 | 1.82 |
ENSRNOT00000075840
ENSRNOT00000077026 |
Zic3
|
Zic family member 3 |
| chr5_-_150520884 | 1.80 |
ENSRNOT00000085482
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr2_-_189818224 | 1.78 |
ENSRNOT00000020575
|
Ints3
|
integrator complex subunit 3 |
| chr3_+_150188455 | 1.76 |
ENSRNOT00000072917
|
AABR07054370.1
|
|
| chr2_+_187740531 | 1.75 |
ENSRNOT00000092653
|
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr1_+_184301493 | 1.74 |
ENSRNOT00000078096
|
Insc
|
inscuteable homolog (Drosophila) |
| chr20_+_28815039 | 1.72 |
ENSRNOT00000075800
|
Sowahc
|
sosondowah ankyrin repeat domain family member C |
| chr1_-_77844189 | 1.60 |
ENSRNOT00000017555
|
Gltscr2
|
glioma tumor suppressor candidate region gene 2 |
| chr6_+_110093819 | 1.59 |
ENSRNOT00000013602
|
Gpatch2l
|
G patch domain containing 2-like |
| chr1_+_101055622 | 1.55 |
ENSRNOT00000079189
|
Nosip
|
nitric oxide synthase interacting protein |
| chr2_-_88414012 | 1.49 |
ENSRNOT00000014762
|
Lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr5_-_76756140 | 1.48 |
ENSRNOT00000022107
ENSRNOT00000089251 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr8_-_96203677 | 1.46 |
ENSRNOT00000074515
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr10_+_56605140 | 1.38 |
ENSRNOT00000024079
|
Phf23
|
PHD finger protein 23 |
| chr18_+_14756684 | 1.32 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr8_+_117836701 | 1.31 |
ENSRNOT00000043345
|
Plxnb1
|
plexin B1 |
| chr18_-_69723869 | 1.19 |
ENSRNOT00000000125
|
Elac1
|
elaC ribonuclease Z 1 |
| chr7_-_14302552 | 1.16 |
ENSRNOT00000091368
|
Brd4
|
bromodomain containing 4 |
| chr15_-_108898703 | 1.13 |
ENSRNOT00000067577
|
Zic5
|
Zic family member 5 |
| chr5_+_133864798 | 1.10 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr2_-_33025271 | 1.09 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chrX_+_43497763 | 1.06 |
ENSRNOT00000005014
|
Prdx4
|
peroxiredoxin 4 |
| chr8_+_109455786 | 0.98 |
ENSRNOT00000039593
|
Msl2
|
male-specific lethal 2 homolog (Drosophila) |
| chr15_-_33333417 | 0.94 |
ENSRNOT00000018982
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr4_-_7281223 | 0.88 |
ENSRNOT00000019666
|
Slc4a2
|
solute carrier family 4 member 2 |
| chr3_+_8450275 | 0.84 |
ENSRNOT00000020073
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr9_+_82700468 | 0.82 |
ENSRNOT00000027227
|
Inha
|
inhibin alpha subunit |
| chr2_+_198823366 | 0.78 |
ENSRNOT00000083769
ENSRNOT00000028814 |
Pias3
|
protein inhibitor of activated STAT, 3 |
| chr17_+_90802393 | 0.78 |
ENSRNOT00000003535
|
Edaradd
|
EDAR-associated death domain |
| chr1_-_174378926 | 0.77 |
ENSRNOT00000018309
|
Tmem9b
|
TMEM9 domain family, member B |
| chr2_+_57206613 | 0.74 |
ENSRNOT00000082694
ENSRNOT00000046069 |
Nup155
|
nucleoporin 155 |
| chr3_-_2431148 | 0.74 |
ENSRNOT00000012978
|
Nelfb
|
negative elongation factor complex member B |
| chr6_+_104291340 | 0.72 |
ENSRNOT00000089313
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr20_+_2043719 | 0.67 |
ENSRNOT00000030174
|
RT1-M4
|
RT1 class Ib, locus M4 |
| chr3_+_8450612 | 0.66 |
ENSRNOT00000040457
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr11_+_71151132 | 0.64 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr5_+_104941066 | 0.63 |
ENSRNOT00000009225
|
Rraga
|
Ras-related GTP binding A |
| chr8_-_96266342 | 0.57 |
ENSRNOT00000078891
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr3_+_112916217 | 0.56 |
ENSRNOT00000030065
|
Tmem62
|
transmembrane protein 62 |
| chr1_+_218945508 | 0.56 |
ENSRNOT00000034240
|
RGD1311946
|
similar to RIKEN cDNA 1810055G02 |
| chr7_+_117055395 | 0.47 |
ENSRNOT00000091960
|
Mapk15
|
mitogen-activated protein kinase 15 |
| chr9_+_6966908 | 0.46 |
ENSRNOT00000072796
|
St6gal2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
| chr15_+_33333420 | 0.44 |
ENSRNOT00000049927
|
RGD1308430
|
similar to 1700123O20Rik protein |
| chr5_-_79899054 | 0.40 |
ENSRNOT00000074379
|
AC229945.1
|
|
| chr10_-_15211325 | 0.40 |
ENSRNOT00000027083
|
Rhot2
|
ras homolog family member T2 |
| chr10_+_43446526 | 0.36 |
ENSRNOT00000036959
|
Larp1
|
La ribonucleoprotein domain family, member 1 |
| chr20_-_5441706 | 0.31 |
ENSRNOT00000000549
|
Vps52
|
VPS52 GARP complex subunit |
| chr7_+_123222161 | 0.26 |
ENSRNOT00000007357
|
RGD1306782
|
similar to RIKEN cDNA 1700029P11 |
| chr14_+_78939903 | 0.19 |
ENSRNOT00000078562
ENSRNOT00000088221 |
Man2b2
Mrfap1
|
mannosidase, alpha, class 2B, member 2 Morf4 family associated protein 1 |
| chr6_+_24064738 | 0.11 |
ENSRNOT00000035362
|
Ypel5
|
yippee-like 5 |
| chr3_-_5976244 | 0.11 |
ENSRNOT00000009893
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr14_-_86706626 | 0.10 |
ENSRNOT00000082893
|
H2afv
|
H2A histone family, member V |
| chr10_-_105182834 | 0.09 |
ENSRNOT00000068050
|
Exoc7
|
exocyst complex component 7 |
| chr14_+_83560541 | 0.07 |
ENSRNOT00000057738
ENSRNOT00000085228 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr2_-_154542919 | 0.06 |
ENSRNOT00000076880
|
Slc33a1
|
solute carrier family 33 member 1 |
| chr5_+_140870140 | 0.05 |
ENSRNOT00000074347
|
Hpcal4
|
hippocalcin-like 4 |
| chr11_+_86890585 | 0.01 |
ENSRNOT00000002579
|
Ranbp1
|
RAN binding protein 1 |
| chr3_+_9366053 | 0.00 |
ENSRNOT00000073999
|
Fibcd1l1
|
fibrinogen C domain containing 1-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ep300
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 3.1 | 12.3 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 2.7 | 10.6 | GO:0030221 | basophil differentiation(GO:0030221) |
| 2.0 | 6.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 1.7 | 5.1 | GO:0021530 | neuroblast differentiation(GO:0014016) spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) medulla oblongata development(GO:0021550) musculoskeletal movement, spinal reflex action(GO:0050883) olfactory pit development(GO:0060166) |
| 1.3 | 8.0 | GO:0003383 | apical constriction(GO:0003383) |
| 1.3 | 3.9 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 1.1 | 6.6 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 1.1 | 3.2 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) endoplasmic reticulum mannose trimming(GO:1904380) |
| 1.0 | 5.2 | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905031) |
| 1.0 | 5.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 1.0 | 5.8 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.0 | 13.5 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.8 | 5.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.8 | 3.2 | GO:0030578 | PML body organization(GO:0030578) negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.8 | 3.1 | GO:0090309 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.8 | 8.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 4.3 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 0.7 | 9.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.7 | 2.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.6 | 10.6 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.5 | 3.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.5 | 22.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.5 | 14.9 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.5 | 2.8 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.4 | 1.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.4 | 5.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 6.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.4 | 4.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.3 | 2.0 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.3 | 3.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 5.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 1.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 | 0.8 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.3 | 1.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 1.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 4.3 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.2 | 2.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 2.8 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 0.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 0.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 1.8 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 4.2 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 1.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 3.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 1.5 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 0.8 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 0.7 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 1.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 2.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.7 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.4 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 1.7 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 3.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 3.1 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 1.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.3 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 3.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 4.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.4 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 1.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 8.9 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 2.2 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.1 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.1 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.9 | 9.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.9 | 8.0 | GO:0032059 | bleb(GO:0032059) |
| 0.8 | 12.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.7 | 4.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.6 | 4.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.5 | 3.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.4 | 1.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 3.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.4 | 10.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 9.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 3.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 0.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 0.8 | GO:0043511 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin complex(GO:0043511) |
| 0.3 | 2.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) GAIT complex(GO:0097452) |
| 0.2 | 4.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 0.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 1.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 3.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 2.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 5.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 3.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 2.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 2.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 7.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.3 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.1 | 5.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 13.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 5.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 9.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 5.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 1.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 3.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 4.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 4.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 1.6 | 6.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.9 | 2.8 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.9 | 5.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.8 | 5.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.7 | 2.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.6 | 3.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.6 | 12.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.6 | 5.8 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.5 | 22.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.5 | 12.3 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.5 | 9.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.4 | 1.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.4 | 2.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 3.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.4 | 1.2 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.4 | 5.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.4 | 1.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.3 | 13.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 3.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 3.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 3.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 3.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 2.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 5.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 4.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 1.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 8.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.1 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 5.0 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 2.8 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 1.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 3.2 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 7.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 14.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.2 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 2.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 8.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 9.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 6.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 4.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 4.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.5 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 6.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.4 | 22.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 5.8 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.3 | 9.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 12.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 7.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 9.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 3.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 22.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.6 | 9.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 9.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 8.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 3.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 6.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 5.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 2.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 5.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 3.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 5.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 3.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 9.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 10.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 2.5 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 5.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |