Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
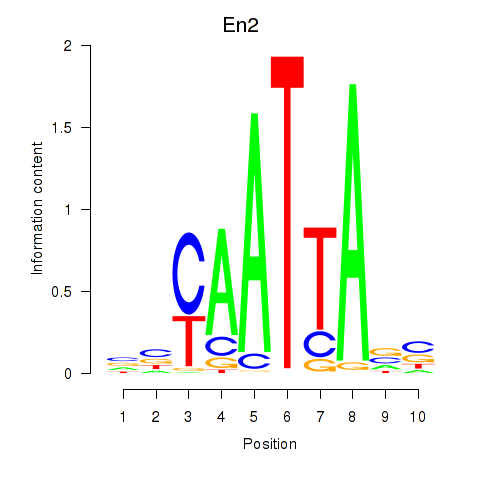
Results for En2
Z-value: 0.42
Transcription factors associated with En2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
En2
|
ENSRNOG00000006846 | engrailed homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| En2 | rn6_v1_chr4_+_438668_438668 | -0.05 | 4.1e-01 | Click! |
Activity profile of En2 motif
Sorted Z-values of En2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_78985990 | 19.91 |
ENSRNOT00000009248
|
Ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr18_+_81694808 | 15.89 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chr2_+_54466280 | 14.29 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr6_-_7058314 | 11.53 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr16_+_10267482 | 9.68 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chrM_+_11736 | 8.52 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr11_-_71284939 | 8.06 |
ENSRNOT00000002421
|
AABR07034428.1
|
|
| chr16_+_50049828 | 6.27 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr11_-_60882379 | 6.05 |
ENSRNOT00000002799
|
Cd200r1
|
CD200 receptor 1 |
| chr6_-_115616766 | 5.92 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr20_-_45260119 | 5.84 |
ENSRNOT00000000718
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr7_-_126382449 | 5.83 |
ENSRNOT00000085540
|
7530416G11Rik
|
RIKEN cDNA 7530416G11 gene |
| chr6_+_73553210 | 5.79 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chrM_+_9451 | 5.55 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr7_+_41475163 | 5.48 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr20_+_44680449 | 5.47 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chrM_+_7006 | 5.42 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr13_+_44424689 | 5.35 |
ENSRNOT00000005206
|
Acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr19_-_17399548 | 5.13 |
ENSRNOT00000075991
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chrM_+_9870 | 5.10 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr18_+_31396922 | 5.07 |
ENSRNOT00000026273
|
RGD735029
|
SEL1 domain containing protein RGD735029 |
| chr6_-_128149220 | 4.89 |
ENSRNOT00000014204
|
Gsc
|
goosecoid homeobox |
| chr20_-_157861 | 4.76 |
ENSRNOT00000084461
|
RT1-CE10
|
RT1 class I, locus CE10 |
| chrM_+_7919 | 4.69 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr4_+_55772627 | 4.63 |
ENSRNOT00000048252
|
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr1_-_164101578 | 4.55 |
ENSRNOT00000022176
|
Uvrag
|
UV radiation resistance associated |
| chr20_-_157665 | 4.49 |
ENSRNOT00000048858
ENSRNOT00000079494 |
RT1-CE10
|
RT1 class I, locus CE10 |
| chr1_-_218810118 | 4.32 |
ENSRNOT00000065950
ENSRNOT00000020886 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr13_+_91054974 | 4.24 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr14_-_2032593 | 4.20 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr4_+_55772377 | 3.94 |
ENSRNOT00000047698
|
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr1_-_185143272 | 3.81 |
ENSRNOT00000027752
|
Nucb2
|
nucleobindin 2 |
| chr19_-_24732024 | 3.70 |
ENSRNOT00000037608
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr7_-_70407177 | 3.70 |
ENSRNOT00000049895
|
Os9
|
OS9, endoplasmic reticulum lectin |
| chr6_+_128750795 | 3.61 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr10_-_87468711 | 3.48 |
ENSRNOT00000039983
|
Krtap3-3
|
keratin associated protein 3-3 |
| chr10_+_5199226 | 3.45 |
ENSRNOT00000003544
|
Dexi
|
dexamethasone-induced transcript |
| chr4_-_100252755 | 3.41 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chrX_+_124321551 | 3.38 |
ENSRNOT00000074486
|
LOC100910807
|
transcriptional regulator Kaiso-like |
| chr2_-_138833933 | 3.36 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr18_-_31343797 | 3.32 |
ENSRNOT00000081850
|
Pcdh1
|
protocadherin 1 |
| chr1_-_101095594 | 3.15 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr6_+_8886591 | 2.94 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr14_+_36216002 | 2.76 |
ENSRNOT00000038506
|
Scfd2
|
sec1 family domain containing 2 |
| chr1_-_276228574 | 2.58 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr3_-_43119159 | 2.55 |
ENSRNOT00000041394
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr4_+_6827429 | 2.52 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr11_-_60679555 | 2.50 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr9_+_49647257 | 2.42 |
ENSRNOT00000021899
|
Mrps9
|
mitochondrial ribosomal protein S9 |
| chr7_-_143228060 | 2.39 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr12_-_45801842 | 2.34 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr1_+_60717386 | 2.29 |
ENSRNOT00000015019
|
Ppp2r1a
|
protein phosphatase 2 scaffold subunit A alpha |
| chr14_+_59611434 | 2.18 |
ENSRNOT00000065366
|
Cckar
|
cholecystokinin A receptor |
| chr6_-_11274932 | 2.17 |
ENSRNOT00000021538
|
Msh2
|
mutS homolog 2 |
| chr17_+_78735598 | 2.17 |
ENSRNOT00000020854
|
Hspa14
|
heat shock protein family A, member 14 |
| chr8_+_71514281 | 2.11 |
ENSRNOT00000022256
|
Ns5atp9
|
NS5A (hepatitis C virus) transactivated protein 9 |
| chr11_+_28780446 | 2.11 |
ENSRNOT00000072546
|
Krtap15-1
|
keratin associated protein 15-1 |
| chr8_+_116318963 | 2.07 |
ENSRNOT00000036556
|
Tusc2
|
tumor suppressor candidate 2 |
| chr3_+_111160205 | 2.06 |
ENSRNOT00000019392
|
Chac1
|
ChaC glutathione-specific gamma-glutamylcyclotransferase 1 |
| chr7_-_49732764 | 2.02 |
ENSRNOT00000006453
|
Myf5
|
myogenic factor 5 |
| chrX_-_26376467 | 1.98 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr12_-_24775891 | 1.96 |
ENSRNOT00000074851
|
Wbscr27
|
Williams Beuren syndrome chromosome region 27 |
| chr11_+_13499164 | 1.96 |
ENSRNOT00000013159
|
LOC680121
|
similar to heat shock protein 8 |
| chr18_+_16544508 | 1.94 |
ENSRNOT00000020601
|
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr9_-_85243001 | 1.91 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr4_-_88684415 | 1.83 |
ENSRNOT00000009001
|
LOC500148
|
similar to 40S ribosomal protein S7 (S8) |
| chr3_+_78086943 | 1.78 |
ENSRNOT00000047425
|
Olr691
|
olfactory receptor 691 |
| chr5_-_7941822 | 1.75 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr10_-_87521514 | 1.70 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr7_+_71157664 | 1.55 |
ENSRNOT00000005919
|
Sdr9c7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr15_-_27590931 | 1.53 |
ENSRNOT00000080898
|
Olr1632
|
olfactory receptor 1632 |
| chr3_+_73235594 | 1.44 |
ENSRNOT00000082357
|
Olr463
|
olfactory receptor 463 |
| chr11_+_47924970 | 1.43 |
ENSRNOT00000060577
|
Zpld1
|
zona pellucida-like domain containing 1 |
| chr2_+_189609800 | 1.43 |
ENSRNOT00000089016
|
Slc39a1
|
solute carrier family 39 member 1 |
| chr14_-_6793558 | 1.41 |
ENSRNOT00000002927
|
Mepe
|
matrix extracellular phosphoglycoprotein |
| chr1_-_64147251 | 1.29 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr4_+_1566448 | 1.26 |
ENSRNOT00000088227
|
Olr1243
|
olfactory receptor 1243 |
| chr5_+_68619202 | 1.25 |
ENSRNOT00000039903
|
Olr848
|
olfactory receptor 848 |
| chr14_+_76732650 | 1.24 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr1_-_230649938 | 1.22 |
ENSRNOT00000017615
|
Olr377
|
olfactory receptor 377 |
| chr4_+_1658278 | 1.21 |
ENSRNOT00000073845
|
Olr1250
|
olfactory receptor 1250 |
| chr2_+_72006099 | 1.19 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr11_-_29710849 | 1.17 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr1_-_219369587 | 1.17 |
ENSRNOT00000000599
|
Aip
|
aryl-hydrocarbon receptor-interacting protein |
| chr6_+_8219385 | 1.10 |
ENSRNOT00000040509
|
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr1_-_103631235 | 1.08 |
ENSRNOT00000072393
|
Mrgprb13
|
MAS-related GPR, member B13 |
| chr7_-_143967484 | 1.08 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr3_-_73475608 | 1.08 |
ENSRNOT00000049630
|
Olr480
|
olfactory receptor 480 |
| chr7_+_17956631 | 0.99 |
ENSRNOT00000064298
|
Vom1r106
|
vomeronasal 1 receptor 106 |
| chr12_-_47142852 | 0.95 |
ENSRNOT00000001591
|
Pop5
|
POP5 homolog, ribonuclease P/MRP subunit |
| chr3_-_90751055 | 0.95 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr1_+_243662823 | 0.94 |
ENSRNOT00000067372
|
Dmrt2
|
doublesex and mab-3 related transcription factor 2 |
| chr2_+_66940057 | 0.87 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr1_-_19376301 | 0.85 |
ENSRNOT00000015547
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr8_+_42663633 | 0.84 |
ENSRNOT00000048537
|
Olr1266
|
olfactory receptor 1266 |
| chr2_+_198417619 | 0.81 |
ENSRNOT00000085945
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr5_+_154668179 | 0.77 |
ENSRNOT00000015971
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr3_-_77605962 | 0.77 |
ENSRNOT00000090062
|
Olr665
|
olfactory receptor 665 |
| chr5_+_137255924 | 0.76 |
ENSRNOT00000088022
ENSRNOT00000092755 |
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr2_-_210088949 | 0.74 |
ENSRNOT00000070994
|
Rbm15
|
RNA binding motif protein 15 |
| chr7_-_143497108 | 0.72 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr14_-_43694584 | 0.71 |
ENSRNOT00000041866
|
AABR07072400.1
|
|
| chr9_+_15513063 | 0.69 |
ENSRNOT00000020499
|
Taf8
|
TATA-box binding protein associated factor 8 |
| chr3_-_78790691 | 0.69 |
ENSRNOT00000008750
|
Olr724
|
olfactory receptor 724 |
| chr2_+_51672722 | 0.65 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr4_+_10823281 | 0.64 |
ENSRNOT00000066739
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr1_-_247821728 | 0.62 |
ENSRNOT00000021958
|
Ermp1
|
endoplasmic reticulum metallopeptidase 1 |
| chr7_+_2752680 | 0.61 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr1_+_13595295 | 0.61 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr2_+_266315036 | 0.60 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr14_+_43694183 | 0.56 |
ENSRNOT00000046342
|
RGD1563570
|
similar to ribosomal protein S23 |
| chr7_-_6803318 | 0.56 |
ENSRNOT00000084203
|
Olr954
|
olfactory receptor 954 |
| chr15_+_11298478 | 0.52 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chrX_+_78042859 | 0.51 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr2_+_207930796 | 0.47 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr4_-_170740274 | 0.45 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr12_-_54885 | 0.44 |
ENSRNOT00000090447
|
AABR07034833.1
|
|
| chr19_+_25043680 | 0.42 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr8_+_41252456 | 0.39 |
ENSRNOT00000071683
|
Olr1220
|
olfactory receptor 1220 |
| chr3_-_77911561 | 0.36 |
ENSRNOT00000084947
|
Olr679
|
olfactory receptor 679 |
| chr7_+_144865608 | 0.35 |
ENSRNOT00000091596
ENSRNOT00000055285 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr8_+_47674321 | 0.35 |
ENSRNOT00000026170
ENSRNOT00000078776 |
Trim29
|
tripartite motif-containing 29 |
| chr13_+_79378733 | 0.27 |
ENSRNOT00000039221
|
Tnfsf18
|
tumor necrosis factor superfamily member 18 |
| chr11_+_58624198 | 0.27 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr10_+_63677396 | 0.23 |
ENSRNOT00000005100
|
Slc43a2
|
solute carrier family 43 member 2 |
| chr6_-_36876805 | 0.17 |
ENSRNOT00000006482
|
Msgn1
|
mesogenin 1 |
| chr3_-_160630457 | 0.17 |
ENSRNOT00000018420
|
Semg1
|
semenogelin I |
| chr8_+_40410604 | 0.13 |
ENSRNOT00000049392
|
Olr1202
|
olfactory receptor 1202 |
| chr3_-_60765645 | 0.13 |
ENSRNOT00000050513
|
Atf2
|
activating transcription factor 2 |
| chr11_-_70322690 | 0.02 |
ENSRNOT00000002443
|
Heg1
|
heart development protein with EGF-like domains 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of En2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.3 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 2.9 | 11.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 2.7 | 5.4 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 1.9 | 18.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 1.4 | 5.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.3 | 3.8 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 1.2 | 3.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 1.2 | 5.8 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 1.1 | 4.5 | GO:0097680 | maintenance of Golgi location(GO:0051684) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 1.1 | 3.4 | GO:1903595 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) positive regulation of histamine secretion by mast cell(GO:1903595) |
| 1.0 | 2.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.9 | 5.5 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.7 | 4.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.7 | 2.1 | GO:0052572 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.5 | 2.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.5 | 3.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.5 | 2.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.5 | 9.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.5 | 2.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.4 | 2.2 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.4 | 4.9 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.4 | 5.5 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.4 | 4.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.3 | 0.3 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.3 | 2.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.3 | 4.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.3 | 1.2 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 19.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.3 | 5.5 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.3 | 2.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 0.9 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 0.6 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.2 | 9.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 | 3.1 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 5.9 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 0.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 15.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 1.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.2 | 4.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 0.3 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 2.0 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 1.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 8.6 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.1 | 4.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 1.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 2.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 3.0 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 5.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 1.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 1.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 2.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.8 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.5 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.2 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.0 | 0.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 5.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 1.4 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 1.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 2.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 2.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 14.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.0 | 5.9 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 1.9 | 5.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.8 | 3.7 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 1.1 | 3.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.9 | 8.6 | GO:0097433 | dense body(GO:0097433) |
| 0.7 | 2.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.6 | 9.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 1.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.4 | 19.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 5.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 3.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 4.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.3 | 1.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 1.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.0 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.2 | 19.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 9.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 3.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.3 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.1 | 2.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 4.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 4.2 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.8 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 22.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 2.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 11.7 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 4.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 6.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 4.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.3 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 4.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 19.9 | GO:0019862 | IgA binding(GO:0019862) |
| 1.0 | 5.8 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.7 | 2.2 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.7 | 19.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.6 | 5.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.6 | 3.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.6 | 4.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.5 | 4.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 2.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.4 | 11.5 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.4 | 24.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.3 | 5.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 1.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.3 | 3.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.3 | 1.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 9.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 0.8 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 1.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 9.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 2.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 2.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 4.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 5.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 3.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 2.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.8 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 3.7 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 2.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 4.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 3.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 1.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 8.6 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 2.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 4.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 4.3 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.3 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 2.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 7.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 2.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 4.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.2 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 3.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 5.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.9 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 19.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 5.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 16.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 7.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.5 | 18.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 5.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 2.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 15.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 4.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 3.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 5.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 2.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 2.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |