Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
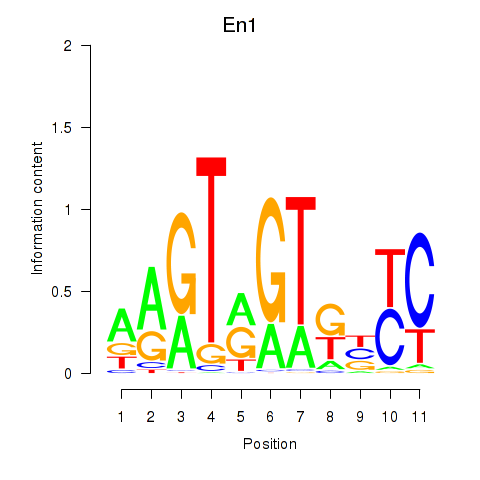
Results for En1
Z-value: 1.31
Transcription factors associated with En1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
En1
|
ENSRNOG00000056580 | engrailed 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| En1 | rn6_v1_chr13_+_36532758_36532758 | -0.08 | 1.6e-01 | Click! |
Activity profile of En1 motif
Sorted Z-values of En1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_54619129 | 53.74 |
ENSRNOT00000059924
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr8_+_49282460 | 37.85 |
ENSRNOT00000021488
|
Cd3d
|
CD3d molecule |
| chr1_-_227441442 | 35.16 |
ENSRNOT00000028433
|
Ms4a1
|
membrane spanning 4-domains A1 |
| chr13_-_77896697 | 33.28 |
ENSRNOT00000003452
|
Tnn
|
tenascin N |
| chr14_+_38030189 | 29.52 |
ENSRNOT00000035623
|
Txk
|
TXK tyrosine kinase |
| chr3_+_16817051 | 29.37 |
ENSRNOT00000071666
|
AABR07051550.1
|
|
| chr4_+_69384145 | 28.69 |
ENSRNOT00000084834
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr15_+_32614002 | 27.60 |
ENSRNOT00000072962
|
AABR07017902.1
|
|
| chr1_-_226791773 | 26.26 |
ENSRNOT00000082482
ENSRNOT00000065376 ENSRNOT00000054812 ENSRNOT00000086669 |
LOC100911215
|
T-cell surface glycoprotein CD5-like |
| chr3_+_17107861 | 25.91 |
ENSRNOT00000043097
|
AABR07051563.1
|
|
| chr10_+_19366793 | 25.15 |
ENSRNOT00000050610
|
Fam196b
|
family with sequence similarity 196, member B |
| chr8_-_117932518 | 24.45 |
ENSRNOT00000028130
|
Camp
|
cathelicidin antimicrobial peptide |
| chrX_-_71168612 | 23.33 |
ENSRNOT00000075934
|
Il2rg
|
interleukin 2 receptor subunit gamma |
| chr2_+_223121410 | 22.71 |
ENSRNOT00000087559
|
AABR07013116.1
|
|
| chr4_+_109467272 | 22.71 |
ENSRNOT00000008212
|
Reg3b
|
regenerating family member 3 beta |
| chr1_-_89329185 | 22.70 |
ENSRNOT00000078284
|
Cd22
|
CD22 molecule |
| chr9_+_2202511 | 22.65 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chrX_+_78372257 | 22.42 |
ENSRNOT00000046164
|
Gpr174
|
G protein-coupled receptor 174 |
| chr13_-_90022269 | 22.11 |
ENSRNOT00000035498
|
Ly9
|
lymphocyte antigen 9 |
| chr3_+_20007192 | 22.01 |
ENSRNOT00000075229
|
AABR07051716.1
|
|
| chr6_+_139405966 | 21.78 |
ENSRNOT00000088974
|
AABR07065693.3
|
|
| chrX_+_134979646 | 21.40 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chrX_+_96991658 | 21.09 |
ENSRNOT00000049969
|
AABR07040288.1
|
|
| chr6_-_138632159 | 20.86 |
ENSRNOT00000082921
ENSRNOT00000040702 |
Ighm
|
immunoglobulin heavy constant mu |
| chr18_+_55466373 | 20.25 |
ENSRNOT00000074629
|
LOC102555392
|
interferon-inducible GTPase 1-like |
| chr15_+_33271015 | 19.94 |
ENSRNOT00000073767
|
Psmb11
|
proteasome subunit beta 11 |
| chr15_+_32809069 | 19.80 |
ENSRNOT00000070848
|
AABR07017902.1
|
|
| chr1_-_100537377 | 19.63 |
ENSRNOT00000026599
|
Spib
|
Spi-B transcription factor |
| chr20_-_4070721 | 19.40 |
ENSRNOT00000000523
|
RT1-Ba
|
RT1 class II, locus Ba |
| chr1_+_86938138 | 19.29 |
ENSRNOT00000075601
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr4_-_103369224 | 18.50 |
ENSRNOT00000075709
|
AABR07061057.1
|
|
| chr4_-_157750088 | 17.98 |
ENSRNOT00000038023
|
Cd27
|
CD27 molecule |
| chr6_+_139177200 | 17.86 |
ENSRNOT00000084131
|
AABR07065676.1
|
|
| chr2_-_112802073 | 17.39 |
ENSRNOT00000093081
|
Ect2
|
epithelial cell transforming 2 |
| chr4_-_103646906 | 17.14 |
ENSRNOT00000047620
|
AABR07061078.1
|
|
| chr3_-_16537433 | 16.41 |
ENSRNOT00000048523
|
AABR07051533.2
|
|
| chr9_-_9675110 | 16.13 |
ENSRNOT00000073294
|
Vav1
|
vav guanine nucleotide exchange factor 1 |
| chr18_+_55391388 | 16.06 |
ENSRNOT00000071612
|
LOC100910934
|
interferon-inducible GTPase 1-like |
| chr4_+_179398621 | 16.04 |
ENSRNOT00000049474
ENSRNOT00000067506 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr4_-_164211819 | 16.02 |
ENSRNOT00000084796
|
LOC497796
|
hypothetical protein LOC497796 |
| chr15_-_37409668 | 15.96 |
ENSRNOT00000011742
|
Gjb6
|
gap junction protein, beta 6 |
| chr5_+_146294030 | 15.68 |
ENSRNOT00000073637
|
AABR07049960.1
|
|
| chr4_-_163403653 | 15.31 |
ENSRNOT00000088151
|
Klrk1
|
killer cell lectin like receptor K1 |
| chr10_+_94566928 | 15.14 |
ENSRNOT00000078446
|
Prr29
|
proline rich 29 |
| chr17_-_43807540 | 14.62 |
ENSRNOT00000074763
|
LOC684762
|
similar to CG31613-PA |
| chr20_+_7279820 | 13.98 |
ENSRNOT00000090820
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr4_+_101882994 | 13.94 |
ENSRNOT00000087773
|
AABR07060963.1
|
|
| chr2_+_219628695 | 13.84 |
ENSRNOT00000067324
|
Sass6
|
SAS-6 centriolar assembly protein |
| chr17_+_78764506 | 13.80 |
ENSRNOT00000077953
|
Suv39h2
|
suppressor of variegation 3-9 homolog 2 |
| chr2_-_216348194 | 13.79 |
ENSRNOT00000087839
|
LOC103689940
|
pancreatic alpha-amylase-like |
| chr6_+_139486775 | 13.75 |
ENSRNOT00000077771
|
AABR07065699.3
|
|
| chr3_-_148057523 | 13.68 |
ENSRNOT00000055408
|
Defb24
|
defensin beta 24 |
| chr3_+_16413080 | 13.43 |
ENSRNOT00000040386
|
LOC100912707
|
Ig kappa chain V19-17-like |
| chr10_-_67401836 | 13.18 |
ENSRNOT00000073071
|
Crlf3
|
cytokine receptor-like factor 3 |
| chr20_-_9876008 | 13.00 |
ENSRNOT00000001537
|
Tff2
|
trefoil factor 2 |
| chr10_-_89454681 | 12.96 |
ENSRNOT00000028109
|
Brca1
|
BRCA1, DNA repair associated |
| chr6_-_138948583 | 12.87 |
ENSRNOT00000084614
|
AABR07065656.2
|
|
| chr8_-_122532070 | 12.81 |
ENSRNOT00000013705
|
Ccr4
|
C-C motif chemokine receptor 4 |
| chr4_-_103115522 | 12.79 |
ENSRNOT00000074206
|
AABR07061044.1
|
|
| chr18_+_55505993 | 12.76 |
ENSRNOT00000043736
|
RGD1309362
|
similar to interferon-inducible GTPase |
| chr15_+_32355565 | 12.76 |
ENSRNOT00000072382
|
AABR07017900.1
|
|
| chr1_+_141120166 | 12.73 |
ENSRNOT00000050759
|
Fanci
|
Fanconi anemia, complementation group I |
| chr17_-_43627629 | 12.61 |
ENSRNOT00000022965
|
Hist1h2af
|
histone cluster 1, H2af |
| chr8_+_96551245 | 12.57 |
ENSRNOT00000039850
|
Bcl2a1
|
BCL2-related protein A1 |
| chr13_-_110257367 | 12.54 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr17_+_43627930 | 12.45 |
ENSRNOT00000081719
|
LOC102549061
|
histone H2B type 1-N-like |
| chr3_+_19690016 | 12.41 |
ENSRNOT00000085460
|
AABR07051707.1
|
|
| chr13_-_74520634 | 12.41 |
ENSRNOT00000077169
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr6_+_122254841 | 11.87 |
ENSRNOT00000005055
|
Gpr65
|
G-protein coupled receptor 65 |
| chr15_+_29208606 | 11.83 |
ENSRNOT00000073722
ENSRNOT00000086054 |
LOC100911980
|
uncharacterized LOC100911980 |
| chr6_-_143131118 | 11.68 |
ENSRNOT00000074930
|
AABR07065834.1
|
|
| chr14_+_76732650 | 11.50 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr15_+_31097291 | 11.47 |
ENSRNOT00000075456
|
AABR07017789.1
|
|
| chr11_+_84745904 | 11.32 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr3_-_172537877 | 11.31 |
ENSRNOT00000072069
|
Ctsz
|
cathepsin Z |
| chr5_+_68717519 | 10.91 |
ENSRNOT00000066226
|
Smc2
|
structural maintenance of chromosomes 2 |
| chr4_-_162025090 | 10.90 |
ENSRNOT00000085887
ENSRNOT00000009904 |
Klrb1a
|
killer cell lectin-like receptor subfamily B, member 1A |
| chr1_+_215609036 | 10.72 |
ENSRNOT00000076187
|
Tnni2
|
troponin I2, fast skeletal type |
| chr3_+_122816924 | 10.68 |
ENSRNOT00000045604
|
RGD1561317
|
similar to ribosomal protein L31 |
| chr18_+_55463308 | 10.61 |
ENSRNOT00000073388
|
LOC100910979
|
interferon-inducible GTPase 1-like |
| chr4_-_157304653 | 10.47 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr6_-_138631997 | 10.47 |
ENSRNOT00000073304
|
AABR07065651.3
|
|
| chr3_+_18805220 | 10.41 |
ENSRNOT00000071216
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr15_+_31618297 | 10.31 |
ENSRNOT00000072332
|
AABR07017831.1
|
|
| chr15_-_29532988 | 10.26 |
ENSRNOT00000074782
|
AABR07017639.1
|
|
| chr7_+_143629455 | 10.20 |
ENSRNOT00000073951
|
Krt18
|
keratin 18 |
| chr16_-_48692476 | 10.12 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr1_+_7726529 | 10.02 |
ENSRNOT00000073460
|
Adat2
|
adenosine deaminase, tRNA-specific 2 |
| chr20_+_3149114 | 9.95 |
ENSRNOT00000084770
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr18_+_44716226 | 9.89 |
ENSRNOT00000086431
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr7_-_142348232 | 9.89 |
ENSRNOT00000078848
|
Galnt6
|
polypeptide N-acetylgalactosaminyltransferase 6 |
| chr1_-_169355117 | 9.88 |
ENSRNOT00000022992
|
RGD1310717
|
similar to RIKEN cDNA E030002O03 |
| chr13_-_77821312 | 9.86 |
ENSRNOT00000082110
|
AABR07021544.1
|
|
| chr2_-_196323881 | 9.68 |
ENSRNOT00000028642
|
Tnfaip8l2
|
TNF alpha induced protein 8 like 2 |
| chr4_-_22192474 | 9.52 |
ENSRNOT00000043822
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr7_-_144880092 | 9.47 |
ENSRNOT00000055281
|
Nfe2
|
nuclear factor, erythroid 2 |
| chr7_+_28654733 | 9.43 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr15_+_28128078 | 9.34 |
ENSRNOT00000082938
|
LOC103694853
|
ribonuclease pancreatic gamma-type |
| chr15_-_35394792 | 9.32 |
ENSRNOT00000028058
|
Gzmc
|
granzyme C |
| chr10_+_44271587 | 9.31 |
ENSRNOT00000047700
|
Trim58
|
tripartite motif-containing 58 |
| chr9_-_60923706 | 9.17 |
ENSRNOT00000047517
|
AABR07067749.1
|
|
| chr2_-_183128932 | 9.17 |
ENSRNOT00000031179
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr5_+_56797502 | 9.16 |
ENSRNOT00000067992
|
Tmem215
|
transmembrane protein 215 |
| chr4_-_78879294 | 9.08 |
ENSRNOT00000084543
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr20_+_3148665 | 9.07 |
ENSRNOT00000086026
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr15_+_4209703 | 9.07 |
ENSRNOT00000082236
|
Ppp3cb
|
protein phosphatase 3 catalytic subunit beta |
| chr1_+_226252226 | 8.98 |
ENSRNOT00000039369
|
AABR07006258.1
|
|
| chr12_-_19314016 | 8.94 |
ENSRNOT00000001825
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr3_-_123631109 | 8.93 |
ENSRNOT00000091493
|
Siglec1
|
sialic acid binding Ig like lectin 1 |
| chr11_+_60383431 | 8.92 |
ENSRNOT00000093295
|
Cd200
|
Cd200 molecule |
| chr1_+_226897625 | 8.79 |
ENSRNOT00000029443
|
Slc15a3
|
solute carrier family 15 member 3 |
| chr10_-_88152064 | 8.74 |
ENSRNOT00000019477
|
Krt16
|
keratin 16 |
| chr4_-_131694755 | 8.61 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr2_+_212435728 | 8.50 |
ENSRNOT00000046960
|
AABR07012868.1
|
|
| chr18_+_25163561 | 8.47 |
ENSRNOT00000042287
ENSRNOT00000049898 ENSRNOT00000017573 |
Bin1
|
bridging integrator 1 |
| chr13_-_61070599 | 8.47 |
ENSRNOT00000005251
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr1_+_72692105 | 8.45 |
ENSRNOT00000032439
|
Tmem150b
|
transmembrane protein 150B |
| chr6_-_138954741 | 8.44 |
ENSRNOT00000083278
|
AABR07065656.7
|
|
| chr13_-_31753800 | 8.44 |
ENSRNOT00000050715
|
AC109707.1
|
|
| chr11_-_88038518 | 8.40 |
ENSRNOT00000051991
|
Rimbp3
|
RIMS binding protein 3 |
| chr10_-_90415070 | 8.39 |
ENSRNOT00000055179
|
Itga2b
|
integrin subunit alpha 2b |
| chr15_-_24056073 | 8.38 |
ENSRNOT00000015100
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr8_-_104155775 | 8.36 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr6_+_139428999 | 8.32 |
ENSRNOT00000084482
|
AABR07065693.2
|
|
| chr13_-_90839411 | 8.30 |
ENSRNOT00000010594
|
Slamf9
|
SLAM family member 9 |
| chr11_+_73936750 | 8.19 |
ENSRNOT00000002350
|
Atp13a3
|
ATPase 13A3 |
| chr15_+_87704340 | 7.92 |
ENSRNOT00000034987
|
Scel
|
sciellin |
| chr15_-_29712639 | 7.90 |
ENSRNOT00000073230
|
AABR07017656.1
|
|
| chr14_-_18849258 | 7.84 |
ENSRNOT00000033406
|
Pf4
|
platelet factor 4 |
| chr7_-_96464049 | 7.81 |
ENSRNOT00000006517
|
Has2
|
hyaluronan synthase 2 |
| chr5_-_78393143 | 7.75 |
ENSRNOT00000043610
|
LOC100910017
|
60S ribosomal protein L31-like |
| chr9_-_61418679 | 7.74 |
ENSRNOT00000078800
|
Ankrd44
|
ankyrin repeat domain 44 |
| chr3_+_58022238 | 7.66 |
ENSRNOT00000002085
|
Hat1
|
histone acetyltransferase 1 |
| chr13_-_53137253 | 7.61 |
ENSRNOT00000011368
|
Gpr25
|
G protein-coupled receptor 25 |
| chrX_-_156891038 | 7.56 |
ENSRNOT00000091495
|
Avpr2
|
arginine vasopressin receptor 2 |
| chr11_+_47146308 | 7.55 |
ENSRNOT00000002191
|
Cep97
|
centrosomal protein 97 |
| chr20_-_5076539 | 7.49 |
ENSRNOT00000076253
ENSRNOT00000037613 |
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr4_+_145489869 | 7.48 |
ENSRNOT00000082618
|
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr6_-_124876710 | 7.35 |
ENSRNOT00000073530
|
Gpr68
|
G protein-coupled receptor 68 |
| chr4_+_78320190 | 7.34 |
ENSRNOT00000032742
ENSRNOT00000091359 |
Gimap4
|
GTPase, IMAP family member 4 |
| chr5_-_153924896 | 7.29 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr4_+_156051915 | 7.29 |
ENSRNOT00000087933
ENSRNOT00000077840 |
Clec4a
|
C-type lectin domain family 4, member A |
| chr4_+_102290338 | 7.22 |
ENSRNOT00000011067
|
AABR07060992.1
|
|
| chr13_+_92136290 | 7.21 |
ENSRNOT00000049819
|
Olr1587
|
olfactory receptor 1587 |
| chr11_+_85633243 | 7.10 |
ENSRNOT00000045807
|
LOC682352
|
Ig lambda chain V-VI region AR-like |
| chrX_-_156891213 | 7.10 |
ENSRNOT00000082899
|
Avpr2
|
arginine vasopressin receptor 2 |
| chr8_-_21492801 | 7.10 |
ENSRNOT00000077465
|
Zfp426
|
zinc finger protein 426 |
| chr5_+_90818736 | 7.09 |
ENSRNOT00000093480
ENSRNOT00000009083 |
Kdm4c
|
lysine demethylase 4C |
| chr1_+_212695735 | 7.02 |
ENSRNOT00000049269
|
Cd163l1
|
CD163 molecule-like 1 |
| chr10_+_29165577 | 7.01 |
ENSRNOT00000078415
|
Ccnjl
|
cyclin J-like |
| chr7_+_14643704 | 6.99 |
ENSRNOT00000044642
|
AABR07055875.1
|
|
| chr3_-_161246351 | 6.95 |
ENSRNOT00000020348
|
Tnnc2
|
troponin C2, fast skeletal type |
| chr5_+_131915382 | 6.88 |
ENSRNOT00000087012
|
Skint1
|
selection and upkeep of intraepithelial T cells 1 |
| chr5_-_83648044 | 6.84 |
ENSRNOT00000046725
|
RGD1561195
|
similar to ribosomal protein L31 |
| chr16_-_54137660 | 6.81 |
ENSRNOT00000085435
|
Pcm1
|
pericentriolar material 1 |
| chrX_-_115426083 | 6.76 |
ENSRNOT00000014756
|
AABR07040947.1
|
|
| chr5_+_139007642 | 6.69 |
ENSRNOT00000066774
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr13_-_89306219 | 6.66 |
ENSRNOT00000004183
ENSRNOT00000079247 |
Fcrla
|
Fc receptor-like A |
| chr7_-_2961873 | 6.63 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr2_+_198040536 | 6.60 |
ENSRNOT00000028744
|
Anp32e
|
acidic nuclear phosphoprotein 32 family member E |
| chr6_+_127927650 | 6.56 |
ENSRNOT00000057271
|
LOC100909605
|
serine protease inhibitor A3F-like |
| chr3_+_79498179 | 6.50 |
ENSRNOT00000030750
|
Nup160
|
nucleoporin 160 |
| chr4_+_45567573 | 6.46 |
ENSRNOT00000089824
|
Ankrd7
|
ankyrin repeat domain 7 |
| chr4_+_71652354 | 6.41 |
ENSRNOT00000022672
ENSRNOT00000022543 |
Casp2
|
caspase 2 |
| chr18_-_32670665 | 6.40 |
ENSRNOT00000019409
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr3_+_103753238 | 6.36 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr8_+_52729003 | 6.35 |
ENSRNOT00000081738
|
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr8_+_22050222 | 6.27 |
ENSRNOT00000028096
|
Icam5
|
intercellular adhesion molecule 5 |
| chr6_-_125317593 | 6.10 |
ENSRNOT00000074142
|
LOC102553138
|
ovarian cancer G-protein coupled receptor 1-like |
| chr10_-_34242985 | 6.06 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chrX_-_1346181 | 6.04 |
ENSRNOT00000042736
|
LOC102555453
|
60S ribosomal protein L12-like |
| chr9_-_46309451 | 6.01 |
ENSRNOT00000018684
|
Rnf149
|
ring finger protein 149 |
| chr7_-_59763219 | 5.98 |
ENSRNOT00000041210
|
RGD1564883
|
similar to 60S ribosomal protein L12 |
| chr10_+_43817947 | 5.98 |
ENSRNOT00000032816
|
Lypd8
|
LY6/PLAUR domain containing 8 |
| chr15_-_52399074 | 5.90 |
ENSRNOT00000018440
|
Xpo7
|
exportin 7 |
| chr17_+_15429708 | 5.88 |
ENSRNOT00000093261
|
LOC679342
|
hypothetical protein LOC679342 |
| chr6_-_143768985 | 5.87 |
ENSRNOT00000046694
|
Ighv15-2
|
immunoglobulin heavy variable V15-2 |
| chr10_+_71008709 | 5.86 |
ENSRNOT00000055923
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr4_+_31704747 | 5.83 |
ENSRNOT00000047850
|
AABR07059675.1
|
|
| chr10_-_82399484 | 5.76 |
ENSRNOT00000082972
ENSRNOT00000055582 |
Xylt2
|
xylosyltransferase 2 |
| chr4_+_183896303 | 5.73 |
ENSRNOT00000055437
|
RGD1309621
|
similar to hypothetical protein FLJ10652 |
| chr7_+_27081667 | 5.60 |
ENSRNOT00000066143
|
Nfyb
|
nuclear transcription factor Y subunit beta |
| chr9_-_73948583 | 5.58 |
ENSRNOT00000018097
|
Myl1
|
myosin, light chain 1 |
| chr14_+_104821215 | 5.58 |
ENSRNOT00000007162
|
Sertad2
|
SERTA domain containing 2 |
| chr15_-_51834030 | 5.54 |
ENSRNOT00000024895
|
Ccar2
|
cell cycle and apoptosis regulator 2 |
| chr10_-_53595827 | 5.53 |
ENSRNOT00000004560
|
Adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr4_+_9882904 | 5.53 |
ENSRNOT00000016909
|
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr10_+_104019888 | 5.49 |
ENSRNOT00000032016
|
Slc16a5
|
solute carrier family 16 member 5 |
| chr12_+_2948570 | 5.46 |
ENSRNOT00000071450
|
LOC100363914
|
hypothetical LOC100363914 |
| chr14_+_96300596 | 5.44 |
ENSRNOT00000071967
|
LOC302228
|
similar to Spindlin-like protein 2 (SPIN-2) |
| chr9_+_81656116 | 5.43 |
ENSRNOT00000083421
|
Slc11a1
|
solute carrier family 11 member 1 |
| chr15_-_82581916 | 5.43 |
ENSRNOT00000057891
|
RGD1560069
|
similar to ribosomal protein L27 |
| chr1_-_116567189 | 5.42 |
ENSRNOT00000048996
|
LOC100912195
|
protein BEX1-like |
| chr5_-_120083904 | 5.37 |
ENSRNOT00000064542
|
Jak1
|
Janus kinase 1 |
| chr4_+_98027554 | 5.37 |
ENSRNOT00000009503
|
Serbp1
|
Serpine1 mRNA binding protein 1 |
| chr4_-_183417667 | 5.36 |
ENSRNOT00000089160
|
Fam60a
|
family with sequence similarity 60, member A |
| chr10_+_14122878 | 5.31 |
ENSRNOT00000052008
|
Hs3st6
|
heparan sulfate-glucosamine 3-sulfotransferase 6 |
| chr5_-_138361702 | 5.27 |
ENSRNOT00000067943
ENSRNOT00000064712 |
Ppih
|
peptidylprolyl isomerase H |
| chr10_-_49192693 | 5.26 |
ENSRNOT00000004294
|
Zfp286a
|
zinc finger protein 286A |
| chr20_-_44255064 | 5.16 |
ENSRNOT00000000735
|
Wisp3
|
WNT1 inducible signaling pathway protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of En1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 33.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 7.4 | 29.5 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 6.7 | 53.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 5.7 | 22.7 | GO:1903576 | response to L-arginine(GO:1903576) |
| 5.1 | 15.3 | GO:2000502 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 4.5 | 9.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 4.3 | 13.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 4.1 | 24.4 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 3.2 | 44.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 3.1 | 12.4 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 3.0 | 42.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 2.9 | 8.6 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 2.8 | 8.5 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 2.6 | 23.3 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 2.4 | 14.7 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 2.4 | 21.9 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 2.4 | 9.5 | GO:1905231 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) response to cyclosporin A(GO:1905237) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 2.2 | 22.1 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 2.2 | 10.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 2.1 | 6.4 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 2.0 | 7.8 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 2.0 | 7.8 | GO:1900623 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) positive regulation of hyaluronan biosynthetic process(GO:1900127) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.9 | 9.4 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 1.8 | 7.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 1.8 | 5.4 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 1.8 | 10.6 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 1.8 | 5.3 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 1.7 | 13.8 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 1.6 | 21.4 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 1.6 | 11.3 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 1.6 | 11.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 1.6 | 6.4 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 1.6 | 9.5 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 1.6 | 4.7 | GO:2000769 | regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 1.5 | 7.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.5 | 8.8 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 1.4 | 5.8 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 1.4 | 8.4 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 1.4 | 49.7 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 1.4 | 21.9 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 1.3 | 5.4 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 1.3 | 8.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.2 | 6.1 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) |
| 1.2 | 4.8 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 1.1 | 19.4 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 1.1 | 3.3 | GO:1990743 | protein sialylation(GO:1990743) |
| 1.1 | 17.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.1 | 3.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.0 | 14.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 1.0 | 1.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.9 | 4.6 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.9 | 3.6 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.9 | 2.7 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.9 | 7.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.8 | 11.3 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.8 | 3.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) detection of muscle stretch(GO:0035995) |
| 0.8 | 4.6 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.8 | 7.7 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.7 | 8.5 | GO:0001554 | luteolysis(GO:0001554) |
| 0.7 | 5.5 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.7 | 2.1 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.7 | 2.7 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.7 | 7.4 | GO:0010755 | regulation of plasminogen activation(GO:0010755) positive regulation of plasminogen activation(GO:0010756) |
| 0.7 | 4.0 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.7 | 9.9 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.7 | 3.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.6 | 17.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.6 | 3.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.6 | 1.8 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.6 | 8.9 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.6 | 3.0 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.6 | 8.7 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.6 | 12.8 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.6 | 1.2 | GO:1903660 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.6 | 4.5 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.6 | 6.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.5 | 24.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.5 | 1.6 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.5 | 1.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.5 | 1.6 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.5 | 15.8 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.5 | 10.2 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.5 | 3.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.5 | 7.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.5 | 2.0 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.5 | 3.8 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.5 | 19.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.5 | 3.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.5 | 22.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.5 | 1.4 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.5 | 2.7 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.4 | 8.9 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.4 | 2.7 | GO:0045072 | regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.4 | 3.4 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.4 | 3.0 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.4 | 1.3 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.4 | 4.4 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.4 | 13.2 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.4 | 1.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.4 | 1.6 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.4 | 3.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.4 | 13.8 | GO:0007099 | centriole replication(GO:0007099) |
| 0.4 | 2.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.4 | 13.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.4 | 4.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.4 | 0.7 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.4 | 0.7 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.3 | 6.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.3 | 2.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.3 | 8.7 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.3 | 31.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.3 | 1.6 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.3 | 3.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.3 | 3.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 0.9 | GO:0048749 | compound eye development(GO:0048749) |
| 0.3 | 2.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.3 | 1.8 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.3 | 8.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 0.9 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.3 | 4.9 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.3 | 1.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.3 | 5.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.3 | 2.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 4.0 | GO:0010543 | regulation of platelet activation(GO:0010543) |
| 0.3 | 0.8 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.3 | 2.4 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.3 | 2.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.3 | 1.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 1.5 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.3 | 1.3 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.2 | 6.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 4.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 3.7 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 1.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 2.3 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 9.9 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.2 | 1.6 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 0.4 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.2 | 2.8 | GO:0060013 | righting reflex(GO:0060013) |
| 0.2 | 4.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 16.0 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.2 | 2.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 1.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.2 | 2.0 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 1.5 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.2 | 7.6 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.2 | 8.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 2.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 3.6 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.2 | 1.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.2 | 0.9 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.2 | 10.9 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.2 | 12.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 1.5 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.2 | 0.7 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.2 | 1.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.2 | 2.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 1.9 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 1.4 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.2 | 0.6 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 5.5 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 9.7 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.1 | 1.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 8.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.7 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.9 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 2.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 3.7 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.5 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 2.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 6.1 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 2.5 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 6.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 5.0 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 2.0 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 4.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 5.9 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 3.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 3.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 9.0 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 1.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 1.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 1.8 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.2 | GO:0072126 | positive regulation of glomerular mesangial cell proliferation(GO:0072126) positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:0006863 | purine nucleobase transport(GO:0006863) nucleobase transport(GO:0015851) |
| 0.1 | 6.0 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.5 | GO:0033683 | UV protection(GO:0009650) nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 2.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 4.6 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.1 | 4.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 2.7 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 1.2 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.8 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.1 | 1.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.1 | 8.1 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 2.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 6.1 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.1 | 1.8 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 6.4 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 1.4 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.1 | 5.5 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.9 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 2.5 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 1.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 1.0 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 7.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 1.5 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 14.0 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 2.0 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 0.3 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.8 | GO:0021510 | spinal cord development(GO:0021510) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 91.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 4.3 | 17.4 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 3.2 | 13.0 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 2.8 | 13.8 | GO:0098536 | deuterosome(GO:0098536) |
| 2.5 | 12.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 2.0 | 7.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.9 | 22.5 | GO:0005861 | troponin complex(GO:0005861) |
| 1.8 | 5.5 | GO:0044609 | DBIRD complex(GO:0044609) |
| 1.8 | 5.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 1.8 | 5.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 1.5 | 24.4 | GO:0042581 | specific granule(GO:0042581) |
| 1.5 | 9.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.4 | 10.9 | GO:0000796 | condensin complex(GO:0000796) |
| 1.2 | 19.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.2 | 7.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 1.2 | 4.6 | GO:1990005 | granular vesicle(GO:1990005) |
| 1.0 | 20.4 | GO:0005922 | connexon complex(GO:0005922) |
| 1.0 | 4.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.9 | 3.7 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.9 | 11.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.8 | 8.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.8 | 19.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.8 | 5.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.8 | 8.4 | GO:0002177 | manchette(GO:0002177) |
| 0.7 | 3.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.7 | 6.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.7 | 22.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.7 | 19.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.7 | 9.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.7 | 2.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.6 | 36.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.6 | 7.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.6 | 6.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.6 | 14.0 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.5 | 2.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.5 | 47.5 | GO:0000786 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.5 | 2.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.4 | 2.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.4 | 5.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 4.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 1.8 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.4 | 7.8 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.4 | 1.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.4 | 1.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 110.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.3 | 1.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.3 | 2.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.3 | 7.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 3.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 1.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.3 | 2.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.3 | 1.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 1.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 3.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 2.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 1.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 1.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 18.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 1.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.2 | 2.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 4.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 8.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 17.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 12.2 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 9.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 3.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 9.7 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 1.9 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 7.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 4.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.7 | GO:0042825 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.1 | 1.8 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 6.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 5.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 6.9 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 4.9 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 2.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 3.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 6.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 8.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 12.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 2.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 1.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 2.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 4.3 | 12.8 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 3.9 | 23.3 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 2.9 | 8.8 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 2.3 | 9.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 2.2 | 15.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 2.0 | 13.8 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 1.9 | 5.8 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 1.9 | 9.5 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 1.8 | 5.3 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 1.7 | 36.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 1.6 | 6.4 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 1.6 | 7.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.6 | 7.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.4 | 13.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 1.4 | 22.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 1.4 | 8.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 1.3 | 14.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 1.3 | 7.8 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.2 | 7.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 1.2 | 4.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.1 | 20.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 1.1 | 5.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 1.1 | 5.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 1.1 | 42.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 1.0 | 6.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 1.0 | 5.1 | GO:0070513 | death domain binding(GO:0070513) |
| 1.0 | 12.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.9 | 4.6 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.9 | 27.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.9 | 9.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.9 | 4.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.9 | 5.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.9 | 12.8 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.8 | 5.0 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.8 | 4.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.8 | 3.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.8 | 4.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.8 | 4.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.8 | 15.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.8 | 16.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.8 | 3.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.7 | 5.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.7 | 19.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.7 | 3.4 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.7 | 2.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.6 | 6.4 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.6 | 3.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.6 | 32.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.6 | 2.3 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.6 | 12.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.6 | 5.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.5 | 5.4 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.5 | 21.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.5 | 4.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.5 | 6.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.5 | 2.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.5 | 1.5 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.5 | 9.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.5 | 10.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.5 | 1.4 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.5 | 4.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.4 | 3.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.4 | 15.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.4 | 3.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.4 | 25.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.4 | 2.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.4 | 1.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.4 | 4.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 5.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 2.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 0.7 | GO:0046978 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.3 | 6.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 2.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.3 | 6.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.3 | 9.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.3 | 4.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 13.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.3 | 2.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 2.1 | GO:0003720 | double-stranded telomeric DNA binding(GO:0003691) telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 2.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.3 | 8.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.3 | 2.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.3 | 1.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 5.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 2.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 24.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.2 | 8.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 1.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 7.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 2.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 0.4 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.2 | 13.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 4.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.2 | 5.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 1.0 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.2 | 49.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 2.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 2.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 1.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 3.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 1.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 36.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.2 | 7.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.2 | 3.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 11.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 13.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 39.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 24.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 37.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.5 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 2.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 4.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 10.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 2.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 6.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 2.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.7 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 17.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 7.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 10.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 2.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.7 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 2.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 3.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 2.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.6 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 2.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 3.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 2.1 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0004950 | chemokine receptor activity(GO:0004950) |
| 0.0 | 2.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.2 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 24.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 1.3 | 48.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 1.1 | 44.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 1.0 | 32.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.7 | 35.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.5 | 4.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 22.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.4 | 4.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.4 | 17.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 8.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 5.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 11.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.3 | 17.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.3 | 12.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.3 | 1.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.3 | 6.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.3 | 9.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 3.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 7.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 7.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 8.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.2 | 4.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 3.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 2.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 4.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 4.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 4.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.2 | 6.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 9.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 7.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 5.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 15.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 2.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 37.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 2.4 | 53.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 2.4 | 28.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 1.5 | 23.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 1.2 | 12.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.1 | 20.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.9 | 14.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.8 | 8.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.8 | 31.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.7 | 4.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.7 | 2.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.7 | 2.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.6 | 19.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.5 | 10.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.5 | 11.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 22.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 4.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.4 | 7.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.4 | 4.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 17.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.4 | 3.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 6.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.4 | 6.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.3 | 3.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.3 | 3.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.3 | 7.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 4.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.3 | 3.8 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.3 | 4.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 14.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.3 | 6.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.3 | 4.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 13.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.3 | 1.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 4.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 4.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 3.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 7.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.2 | 5.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 2.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 2.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 2.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 5.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 1.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 1.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 11.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 10.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 2.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 4.0 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.1 | 2.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 5.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 12.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 3.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 5.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 7.4 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.8 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |