Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Emx1_Emx2
Z-value: 0.66
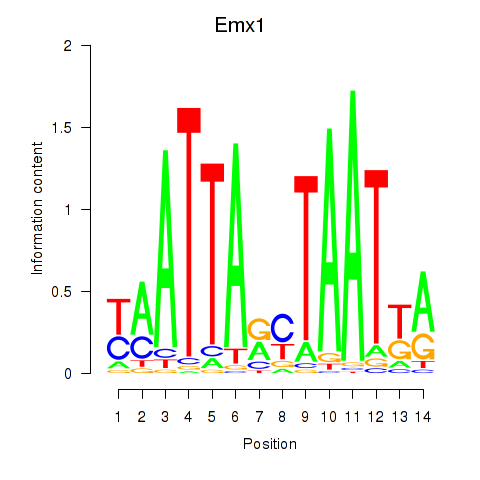
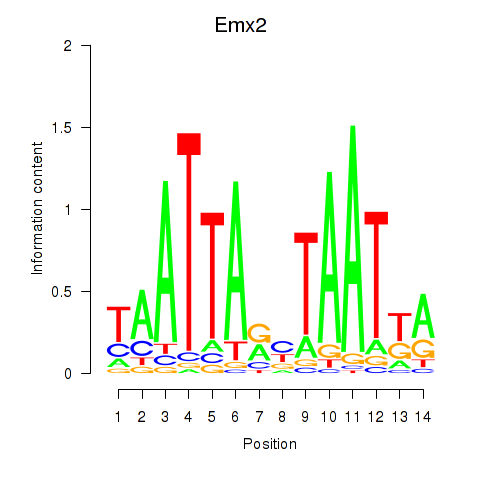
Transcription factors associated with Emx1_Emx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Emx1
|
ENSRNOG00000015493 | empty spiracles homeobox 1 |
|
Emx2
|
ENSRNOG00000009482 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Emx2 | rn6_v1_chr1_+_280633938_280633938 | -0.43 | 5.7e-16 | Click! |
| Emx1 | rn6_v1_chr4_+_116968000_116968000 | -0.11 | 6.0e-02 | Click! |
Activity profile of Emx1_Emx2 motif
Sorted Z-values of Emx1_Emx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_71139267 | 49.19 |
ENSRNOT00000065232
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr1_+_147713892 | 31.14 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr2_-_180914940 | 26.15 |
ENSRNOT00000015732
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr3_+_159368273 | 21.48 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr12_+_47179664 | 18.20 |
ENSRNOT00000001551
|
Cabp1
|
calcium binding protein 1 |
| chr14_+_20266891 | 16.92 |
ENSRNOT00000004174
|
Gc
|
group specific component |
| chr2_+_54466280 | 16.03 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr2_-_216382244 | 15.95 |
ENSRNOT00000086695
ENSRNOT00000087259 |
LOC103689940
|
pancreatic alpha-amylase-like |
| chr1_-_148119857 | 15.35 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr10_+_103206014 | 14.37 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr1_+_248428099 | 14.30 |
ENSRNOT00000050984
|
Mbl2
|
mannose binding lectin 2 |
| chr2_-_216348194 | 13.14 |
ENSRNOT00000087839
|
LOC103689940
|
pancreatic alpha-amylase-like |
| chr15_-_80713153 | 12.42 |
ENSRNOT00000063800
|
Klhl1
|
kelch-like family member 1 |
| chrX_-_14972675 | 10.82 |
ENSRNOT00000079664
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr13_-_76049363 | 10.34 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr4_+_173732248 | 10.32 |
ENSRNOT00000041499
|
Pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr2_-_173668555 | 10.22 |
ENSRNOT00000013452
|
Serpini2
|
serpin family I member 2 |
| chr11_+_80742467 | 9.82 |
ENSRNOT00000002507
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr2_+_52431601 | 9.29 |
ENSRNOT00000023554
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr13_+_47572219 | 9.15 |
ENSRNOT00000088449
ENSRNOT00000087664 ENSRNOT00000005853 |
Pigr
|
polymeric immunoglobulin receptor |
| chr1_+_101603222 | 9.14 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr9_+_73378057 | 8.91 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr5_-_65073012 | 8.83 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr20_+_34258791 | 8.75 |
ENSRNOT00000000468
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr11_+_61605937 | 8.75 |
ENSRNOT00000093455
ENSRNOT00000093242 |
Gramd1c
|
GRAM domain containing 1C |
| chr15_+_28128078 | 8.32 |
ENSRNOT00000082938
|
LOC103694853
|
ribonuclease pancreatic gamma-type |
| chr11_+_57207656 | 8.00 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr10_+_1920529 | 7.83 |
ENSRNOT00000072296
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr10_-_103848035 | 7.46 |
ENSRNOT00000029001
|
Fads6
|
fatty acid desaturase 6 |
| chr4_+_35279063 | 7.40 |
ENSRNOT00000011833
|
Nxph1
|
neurexophilin 1 |
| chr13_+_75177965 | 7.29 |
ENSRNOT00000007321
|
Sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr10_+_64952119 | 7.20 |
ENSRNOT00000012154
|
Pipox
|
pipecolic acid and sarcosine oxidase |
| chr3_-_48831417 | 7.18 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr7_-_50278842 | 6.99 |
ENSRNOT00000088950
|
Syt1
|
synaptotagmin 1 |
| chr6_-_3254779 | 6.98 |
ENSRNOT00000061975
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr7_+_70980422 | 6.87 |
ENSRNOT00000077912
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr3_+_48096954 | 6.82 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr1_-_78180216 | 6.71 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr11_-_782954 | 6.33 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr13_+_71110064 | 6.23 |
ENSRNOT00000088329
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr8_-_113689681 | 6.20 |
ENSRNOT00000056435
|
LOC688828
|
similar to Nucleoside diphosphate-linked moiety X motif 16 (Nudix motif 16) |
| chrX_+_84064427 | 6.11 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr4_-_31730386 | 6.01 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chr11_-_768644 | 5.81 |
ENSRNOT00000090197
|
Epha3
|
Eph receptor A3 |
| chr7_+_27620458 | 5.72 |
ENSRNOT00000059532
|
RGD1560034
|
similar to FLJ25323 protein |
| chr1_+_250426158 | 5.45 |
ENSRNOT00000067643
|
A1cf
|
APOBEC1 complementation factor |
| chr1_+_189233141 | 5.34 |
ENSRNOT00000049380
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr8_+_85503224 | 5.04 |
ENSRNOT00000012348
|
Gsta4
|
glutathione S-transferase alpha 4 |
| chr5_+_22380334 | 4.87 |
ENSRNOT00000009744
|
Clvs1
|
clavesin 1 |
| chr1_-_169321075 | 4.65 |
ENSRNOT00000055216
|
RGD1562433
|
similar to ubiquilin 1 isoform 2 |
| chr11_+_20474483 | 4.52 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr9_+_95161157 | 4.48 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr16_+_39827749 | 4.47 |
ENSRNOT00000074885
|
Wdr17
|
WD repeat domain 17 |
| chr2_-_218658761 | 4.46 |
ENSRNOT00000018318
|
S1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr2_+_220432037 | 4.42 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr5_-_166116516 | 4.41 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr16_+_62153792 | 4.31 |
ENSRNOT00000074543
|
Smim18
|
small integral membrane protein 18 |
| chrX_-_143558521 | 4.30 |
ENSRNOT00000056598
|
LOC688842
|
hypothetical protein LOC688842 |
| chr14_+_48740190 | 4.28 |
ENSRNOT00000031638
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr16_-_64806050 | 4.19 |
ENSRNOT00000015725
|
Dusp26
|
dual specificity phosphatase 26 |
| chr1_-_167911961 | 4.06 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr15_+_12827707 | 4.00 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr9_-_66019065 | 3.88 |
ENSRNOT00000088729
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr9_-_104467973 | 3.88 |
ENSRNOT00000026099
|
Slco6b1
|
solute carrier organic anion transporter family member 6B1 |
| chr2_-_181900856 | 3.80 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr17_-_51912496 | 3.78 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr17_+_25082056 | 3.70 |
ENSRNOT00000037041
|
AABR07027339.1
|
|
| chr9_+_111279495 | 3.64 |
ENSRNOT00000072791
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr2_-_192780631 | 3.58 |
ENSRNOT00000012371
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr1_+_22332090 | 3.52 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr2_+_239415046 | 3.51 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr3_+_2288667 | 3.51 |
ENSRNOT00000012289
|
Entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr2_-_198706428 | 3.35 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr18_-_43945273 | 3.33 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr20_-_45126062 | 3.32 |
ENSRNOT00000000720
|
RGD1310495
|
similar to KIAA1919 protein |
| chr15_+_80040842 | 3.32 |
ENSRNOT00000043065
|
RGD1306441
|
similar to RIKEN cDNA 4921530L21 |
| chr9_+_66058047 | 3.25 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr7_-_76488216 | 3.22 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr8_-_55144087 | 3.19 |
ENSRNOT00000039045
|
Dixdc1
|
DIX domain containing 1 |
| chr4_-_55011415 | 3.19 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr11_-_11585078 | 3.19 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr10_-_34301197 | 3.16 |
ENSRNOT00000044667
|
Olr1383
|
olfactory receptor 1383 |
| chr15_-_33576331 | 3.16 |
ENSRNOT00000021851
|
Slc22a17
|
solute carrier family 22, member 17 |
| chr11_+_7265828 | 3.11 |
ENSRNOT00000084765
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chrX_-_124252447 | 3.01 |
ENSRNOT00000061546
|
Rhox12
|
reproductive homeobox 12 |
| chr6_-_14523961 | 2.95 |
ENSRNOT00000071402
|
Nrxn1
|
neurexin 1 |
| chr6_-_23404368 | 2.94 |
ENSRNOT00000036374
|
Fam179a
|
family with sequence similarity 179, member A |
| chr15_+_5904569 | 2.92 |
ENSRNOT00000072599
|
LOC102546376
|
disks large homolog 5-like |
| chr4_-_68349273 | 2.87 |
ENSRNOT00000016251
|
Prss37
|
protease, serine, 37 |
| chr1_-_22570303 | 2.75 |
ENSRNOT00000035539
|
Taar3
|
trace amine-associated receptor 3 |
| chr3_+_79613171 | 2.73 |
ENSRNOT00000011467
|
Agbl2
|
ATP/GTP binding protein-like 2 |
| chr14_-_106248539 | 2.65 |
ENSRNOT00000067668
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr7_-_145048283 | 2.64 |
ENSRNOT00000055275
|
Gtsf1
|
gametocyte specific factor 1 |
| chr15_-_28044210 | 2.64 |
ENSRNOT00000033809
|
Rnase1l1
|
ribonuclease, RNase A family, 1-like 1 (pancreatic) |
| chrX_-_135041027 | 2.58 |
ENSRNOT00000006484
|
Zdhhc9
|
zinc finger, DHHC-type containing 9 |
| chr1_-_279277339 | 2.55 |
ENSRNOT00000023667
|
Gfra1
|
GDNF family receptor alpha 1 |
| chr2_+_18392142 | 2.54 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr1_-_252116971 | 2.53 |
ENSRNOT00000035013
|
Lipo1
|
lipase, member O1 |
| chr2_-_137153551 | 2.47 |
ENSRNOT00000083443
|
AABR07010476.1
|
|
| chr3_-_143192408 | 2.47 |
ENSRNOT00000006885
|
LOC689081
|
similar to cystatin E2 |
| chr9_-_97290639 | 2.45 |
ENSRNOT00000026491
ENSRNOT00000056724 |
Iqca1
|
IQ motif containing with AAA domain 1 |
| chr3_+_95233874 | 2.41 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chr1_-_189182306 | 2.40 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr7_-_54823956 | 2.39 |
ENSRNOT00000073180
|
Glipr1l2
|
GLI pathogenesis-related 1 like 2 |
| chr9_+_8093262 | 2.39 |
ENSRNOT00000080314
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr16_+_70644474 | 2.34 |
ENSRNOT00000045955
|
LOC100359503
|
ribosomal protein S28-like |
| chr10_-_86393141 | 2.33 |
ENSRNOT00000009485
|
Mien1
|
migration and invasion enhancer 1 |
| chr10_-_61088827 | 2.32 |
ENSRNOT00000047421
|
Olr1517
|
olfactory receptor 1517 |
| chr7_+_144531814 | 2.32 |
ENSRNOT00000033300
|
Hoxc13
|
homeobox C13 |
| chr2_-_173836854 | 2.30 |
ENSRNOT00000074278
|
Wdr49
|
WD repeat domain 49 |
| chrX_+_136466779 | 2.28 |
ENSRNOT00000093268
ENSRNOT00000068717 |
Arhgap36
|
Rho GTPase activating protein 36 |
| chr4_-_58250798 | 2.25 |
ENSRNOT00000048436
|
Klf14
|
Kruppel-like factor 14 |
| chr14_-_21909646 | 2.23 |
ENSRNOT00000088024
|
Csn1s2b
|
casein alpha s2-like B |
| chr20_+_915079 | 2.21 |
ENSRNOT00000074181
|
LOC100910382
|
olfactory receptor 14J1-like |
| chr8_-_3272306 | 2.19 |
ENSRNOT00000040365
|
AABR07068955.1
|
|
| chr3_-_161246351 | 2.19 |
ENSRNOT00000020348
|
Tnnc2
|
troponin C2, fast skeletal type |
| chr6_-_23291568 | 2.14 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr15_+_52265557 | 2.13 |
ENSRNOT00000015969
|
Nudt18
|
nudix hydrolase 18 |
| chr17_-_18590536 | 2.12 |
ENSRNOT00000078992
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr20_+_3176107 | 2.10 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr9_+_23596964 | 2.09 |
ENSRNOT00000064279
|
LOC108351994
|
exocrine gland-secreted peptide 1-like |
| chr19_-_11669578 | 2.08 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr9_+_101197255 | 2.08 |
ENSRNOT00000045660
|
RGD1564236
|
similar to OBOX3 |
| chr1_-_230079916 | 2.07 |
ENSRNOT00000091024
|
Olr358
|
olfactory receptor 358 |
| chr9_+_8054466 | 2.00 |
ENSRNOT00000081513
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr6_-_10515370 | 2.00 |
ENSRNOT00000020845
|
Atp6v1e2
|
ATPase H+ transporting V1 subunit E2 |
| chr2_+_195651930 | 1.97 |
ENSRNOT00000028299
|
Tdrkh
|
tudor and KH domain containing |
| chr5_-_110978644 | 1.94 |
ENSRNOT00000090548
|
AABR07049228.1
|
|
| chr2_-_250923744 | 1.89 |
ENSRNOT00000084996
|
Clca1
|
chloride channel accessory 1 |
| chr3_+_147952879 | 1.88 |
ENSRNOT00000031922
|
Defb20
|
defensin beta 20 |
| chr2_+_52855671 | 1.88 |
ENSRNOT00000080851
|
AABR07008319.1
|
|
| chr10_-_87195075 | 1.86 |
ENSRNOT00000014851
|
Krt24
|
keratin 24 |
| chr8_+_119083900 | 1.85 |
ENSRNOT00000051947
ENSRNOT00000048655 |
Prss44
|
protease, serine, 44 |
| chr15_+_34234755 | 1.85 |
ENSRNOT00000059987
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr6_+_95205153 | 1.81 |
ENSRNOT00000007339
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr4_+_33908299 | 1.81 |
ENSRNOT00000010375
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr1_+_169660076 | 1.80 |
ENSRNOT00000073447
|
Olr160
|
olfactory receptor 160 |
| chr1_+_22319353 | 1.78 |
ENSRNOT00000038523
|
Taar9
|
trace amine-associated receptor 9 |
| chr3_-_74631883 | 1.77 |
ENSRNOT00000013215
|
Olr539
|
olfactory receptor 539 |
| chrX_-_23187341 | 1.76 |
ENSRNOT00000000180
|
Alas2
|
5'-aminolevulinate synthase 2 |
| chr17_+_89951752 | 1.74 |
ENSRNOT00000057572
ENSRNOT00000091023 |
Abi1
|
abl-interactor 1 |
| chr8_+_117170620 | 1.72 |
ENSRNOT00000075271
|
LOC498675
|
hypothetical LOC498675 |
| chr3_-_30930141 | 1.72 |
ENSRNOT00000037001
|
AABR07051982.1
|
|
| chr13_+_88265331 | 1.71 |
ENSRNOT00000031190
|
Ccdc190
|
coiled-coil domain containing 190 |
| chr2_-_132301073 | 1.69 |
ENSRNOT00000058288
|
RGD1563562
|
similar to GTPase activating protein testicular GAP1 |
| chr8_+_36909433 | 1.66 |
ENSRNOT00000074169
|
LOC100359924
|
prostate and testis expressed N-like |
| chr14_-_21810314 | 1.66 |
ENSRNOT00000002671
|
Csn3
|
casein kappa |
| chr15_-_14510828 | 1.65 |
ENSRNOT00000039347
|
Sntn
|
sentan, cilia apical structure protein |
| chr17_-_62299655 | 1.60 |
ENSRNOT00000024855
|
Gjd4
|
gap junction protein, delta 4 |
| chr15_+_28894627 | 1.57 |
ENSRNOT00000017795
|
Olr1643
|
olfactory receptor 1643 |
| chr20_-_3166569 | 1.56 |
ENSRNOT00000091390
|
AABR07044362.1
|
|
| chr4_+_88694583 | 1.55 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr17_-_38858331 | 1.54 |
ENSRNOT00000059738
|
Prl2c1
|
Prolactin family 2, subfamily c, member 1 |
| chr16_-_20873344 | 1.54 |
ENSRNOT00000027381
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr1_+_201913148 | 1.53 |
ENSRNOT00000036128
|
Fam24a
|
family with sequence similarity 24, member A |
| chr2_+_88217188 | 1.52 |
ENSRNOT00000014267
|
Car1
|
carbonic anhydrase I |
| chr2_-_122641409 | 1.52 |
ENSRNOT00000068334
|
Dcun1d1
|
defective in cullin neddylation 1 domain containing 1 |
| chr9_+_10941613 | 1.49 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr1_+_279633671 | 1.48 |
ENSRNOT00000036012
ENSRNOT00000091669 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr20_-_556982 | 1.47 |
ENSRNOT00000047478
|
Olr1683
|
olfactory receptor 1683 |
| chr1_-_103717536 | 1.47 |
ENSRNOT00000029920
|
AABR07003291.1
|
|
| chr1_+_170205591 | 1.45 |
ENSRNOT00000071063
|
LOC686660
|
similar to olfactory receptor 692 |
| chr10_+_110445797 | 1.43 |
ENSRNOT00000054920
|
Narf
|
nuclear prelamin A recognition factor |
| chr10_+_45307007 | 1.42 |
ENSRNOT00000003885
|
Trim17
|
tripartite motif-containing 17 |
| chrX_-_23469041 | 1.39 |
ENSRNOT00000065789
|
LOC685699
|
hypothetical protein LOC685699 |
| chrX_+_144994139 | 1.39 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr1_-_67134827 | 1.38 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr10_-_36402619 | 1.36 |
ENSRNOT00000040346
|
Zfp2
|
zinc finger protein 2 |
| chr2_-_61692487 | 1.36 |
ENSRNOT00000078544
|
LOC499541
|
LRRGT00045 |
| chr1_+_22364551 | 1.34 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr20_-_46305157 | 1.33 |
ENSRNOT00000000340
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr2_+_95320283 | 1.30 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr16_-_75241303 | 1.29 |
ENSRNOT00000058056
|
Defb2
|
defensin beta 2 |
| chr14_+_23166633 | 1.28 |
ENSRNOT00000060109
|
Tmprss11g
|
transmembrane protease, serine 11G |
| chr2_+_58668157 | 1.27 |
ENSRNOT00000081192
|
Ugt3a2
|
UDP glycosyltransferase 3 family, polypeptide A2 |
| chr1_-_141533908 | 1.27 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr20_+_734642 | 1.25 |
ENSRNOT00000073543
|
Olr1688
|
olfactory receptor 1688 |
| chr13_+_26463404 | 1.25 |
ENSRNOT00000061779
|
LOC103690041
|
serpin B12-like |
| chr6_+_101532518 | 1.24 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr9_-_23561890 | 1.22 |
ENSRNOT00000060827
|
LOC102553880
|
uncharacterized LOC102553880 |
| chr1_-_133503194 | 1.21 |
ENSRNOT00000077049
|
Mctp2
|
multiple C2 and transmembrane domain containing 2 |
| chr16_-_21473808 | 1.19 |
ENSRNOT00000066776
|
Zfp964
|
zinc finger protein 964 |
| chr10_+_74959285 | 1.18 |
ENSRNOT00000010296
|
Rnf43
|
ring finger protein 43 |
| chr5_+_167141875 | 1.16 |
ENSRNOT00000089314
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr10_+_44506098 | 1.15 |
ENSRNOT00000078135
|
LOC501698
|
similar to olfactory receptor Olr1448 |
| chr7_-_2623781 | 1.14 |
ENSRNOT00000004173
|
Spryd4
|
SPRY domain containing 4 |
| chr9_-_121972055 | 1.14 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr10_+_87846947 | 1.14 |
ENSRNOT00000077436
|
Krtap9-5
|
keratin associated protein 9-5 |
| chr1_+_170147300 | 1.13 |
ENSRNOT00000048075
|
Olr200
|
olfactory receptor 200 |
| chr17_-_42740021 | 1.13 |
ENSRNOT00000023063
|
Prl3a1
|
Prolactin family 3, subfamily a, member 1 |
| chr14_-_21299068 | 1.11 |
ENSRNOT00000065778
|
Amtn
|
amelotin |
| chr18_+_35384743 | 1.09 |
ENSRNOT00000076143
ENSRNOT00000074593 |
LOC100911797
|
serine protease inhibitor Kazal-type 5-like |
| chr1_+_168341595 | 1.08 |
ENSRNOT00000048411
|
Olr84
|
olfactory receptor 84 |
| chr6_+_137323713 | 1.07 |
ENSRNOT00000029017
|
Pld4
|
phospholipase D family, member 4 |
| chr17_+_59714657 | 1.06 |
ENSRNOT00000035675
|
RGD1560860
|
similar to ankyrin repeat domain 26 |
| chr7_+_64769089 | 1.06 |
ENSRNOT00000088861
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr11_-_71938165 | 1.06 |
ENSRNOT00000037795
|
Cep19
|
centrosomal protein 19 |
| chr14_-_21758788 | 1.05 |
ENSRNOT00000038520
|
2310003L06Rik
|
RIKEN cDNA 2310003L06 gene |
| chr1_+_21613148 | 1.02 |
ENSRNOT00000018695
|
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr1_+_168005316 | 1.02 |
ENSRNOT00000020678
|
Olr51
|
olfactory receptor 51 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Emx1_Emx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 30.3 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 6.2 | 31.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 4.0 | 12.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 3.7 | 26.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 3.0 | 9.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 2.4 | 7.2 | GO:0019474 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 2.2 | 6.7 | GO:0071623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 1.6 | 6.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.5 | 18.2 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 1.4 | 4.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 1.4 | 5.5 | GO:0016554 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) cytidine to uridine editing(GO:0016554) |
| 1.4 | 4.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 1.2 | 2.4 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 1.1 | 5.7 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 1.1 | 3.2 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.0 | 4.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.9 | 3.8 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.9 | 2.6 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.9 | 4.4 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.8 | 16.9 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.8 | 4.2 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.8 | 9.8 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.8 | 7.3 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.8 | 10.8 | GO:0015816 | glycine transport(GO:0015816) |
| 0.7 | 9.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.7 | 2.1 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.7 | 2.1 | GO:0002488 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.6 | 15.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.6 | 3.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.6 | 3.2 | GO:0061364 | negative regulation of negative chemotaxis(GO:0050925) apoptotic process involved in luteolysis(GO:0061364) |
| 0.6 | 6.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.6 | 1.8 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.6 | 3.6 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.6 | 3.9 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.5 | 8.8 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.5 | 9.3 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.5 | 4.5 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 3.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.5 | 8.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.4 | 1.8 | GO:1901423 | response to benzene(GO:1901423) |
| 0.4 | 3.5 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.4 | 3.5 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 1.3 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.4 | 10.3 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.4 | 12.4 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.4 | 2.7 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.4 | 5.7 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.4 | 1.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.3 | 1.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.3 | 2.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 1.6 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.3 | 5.0 | GO:0009635 | response to herbicide(GO:0009635) |
| 0.3 | 3.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 0.9 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.3 | 2.3 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 1.2 | GO:0015755 | fructose transport(GO:0015755) |
| 0.2 | 8.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 3.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 21.7 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.2 | 0.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 2.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 1.0 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 4.5 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 10.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.2 | 16.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 2.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 10.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 1.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 1.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.9 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.6 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.1 | 2.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 4.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 3.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 1.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 2.0 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 3.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 2.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 2.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.2 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 2.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.4 | GO:0046549 | retinal rod cell development(GO:0046548) retinal cone cell development(GO:0046549) |
| 0.0 | 2.1 | GO:0043278 | response to morphine(GO:0043278) |
| 0.0 | 1.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 10.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 2.9 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 7.4 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 1.1 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.0 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.7 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.6 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.5 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.8 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 2.5 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 8.7 | GO:0005975 | carbohydrate metabolic process(GO:0005975) |
| 0.0 | 2.9 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 1.0 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 12.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 16.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.7 | 15.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.3 | 3.8 | GO:0043511 | inhibin complex(GO:0043511) |
| 1.2 | 7.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.9 | 2.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.7 | 2.1 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.6 | 4.0 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.5 | 8.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.5 | 4.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.4 | 8.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.3 | 9.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.3 | 1.0 | GO:0098982 | glycine-gated chloride channel complex(GO:0016935) GABA-ergic synapse(GO:0098982) |
| 0.3 | 2.0 | GO:0071547 | piP-body(GO:0071547) |
| 0.3 | 1.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 0.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 14.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 0.7 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.2 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 3.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 9.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 2.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 12.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 6.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 2.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.8 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 16.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 6.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 2.0 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.1 | 18.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 10.0 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.1 | 16.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 4.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 1.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 1.0 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 3.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 19.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 3.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.7 | 29.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 8.7 | 26.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 3.7 | 11.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 2.9 | 14.3 | GO:0005534 | galactose binding(GO:0005534) |
| 2.8 | 16.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 2.2 | 6.7 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.9 | 9.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.8 | 7.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 1.5 | 46.5 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.4 | 18.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 1.4 | 7.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.4 | 4.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 1.2 | 12.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 1.2 | 6.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.1 | 21.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 1.1 | 4.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.1 | 8.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 1.0 | 3.1 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 1.0 | 9.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.9 | 3.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.9 | 3.6 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.9 | 8.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.8 | 10.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.8 | 3.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.7 | 4.4 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.7 | 2.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.7 | 5.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.6 | 1.8 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.6 | 1.8 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.5 | 2.6 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.5 | 5.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.4 | 4.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.4 | 1.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.4 | 3.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.4 | 8.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 2.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.3 | 2.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 5.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 1.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.3 | 1.0 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 0.3 | 2.2 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.3 | 7.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.3 | 1.2 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.3 | 3.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 3.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 5.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.3 | 3.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.2 | 6.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 1.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 2.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 4.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 3.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 5.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.7 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 3.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 12.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 16.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.2 | 6.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 0.5 | GO:0019961 | interferon binding(GO:0019961) |
| 0.2 | 2.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 1.2 | GO:0070061 | fructose binding(GO:0070061) |
| 0.1 | 1.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 3.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 3.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 2.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 1.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 7.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 8.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.2 | GO:1903135 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) cupric ion binding(GO:1903135) |
| 0.1 | 5.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 4.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 2.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 6.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 7.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 1.3 | GO:0035326 | enhancer binding(GO:0035326) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.3 | 6.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.3 | 7.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 9.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 8.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 3.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 8.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 9.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 20.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 7.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 26.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 2.2 | 24.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.7 | 16.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.5 | 10.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.5 | 7.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.5 | 16.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.5 | 3.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.4 | 7.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 2.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 6.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 10.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 3.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 4.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 6.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 7.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 3.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 2.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 8.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 6.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 5.2 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.9 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |