Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
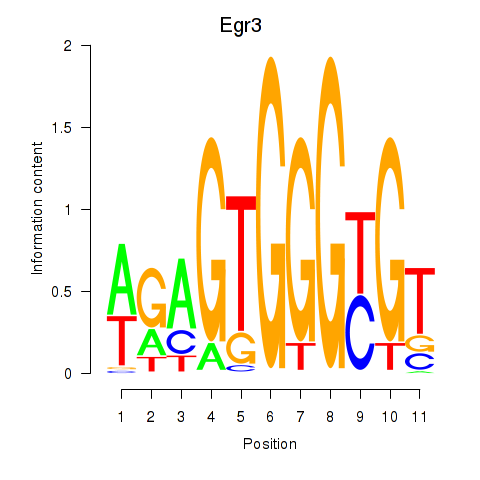
Results for Egr3
Z-value: 0.28
Transcription factors associated with Egr3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr3
|
ENSRNOG00000017828 | early growth response 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr3 | rn6_v1_chr15_+_51756978_51756978 | -0.01 | 8.3e-01 | Click! |
Activity profile of Egr3 motif
Sorted Z-values of Egr3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_180914940 | 17.09 |
ENSRNOT00000015732
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr1_-_256813711 | 12.86 |
ENSRNOT00000021055
|
Rbp4
|
retinol binding protein 4 |
| chr15_+_42653148 | 5.41 |
ENSRNOT00000022095
|
Clu
|
clusterin |
| chr15_+_34187223 | 5.21 |
ENSRNOT00000024978
|
Cpne6
|
copine 6 |
| chr3_-_102151489 | 4.27 |
ENSRNOT00000006349
|
Ano3
|
anoctamin 3 |
| chr10_-_98018014 | 3.45 |
ENSRNOT00000005367
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr13_-_79705705 | 2.69 |
ENSRNOT00000003998
|
Faslg
|
Fas ligand |
| chr1_+_31124825 | 2.48 |
ENSRNOT00000092105
|
AABR07000989.1
|
|
| chr20_-_6500523 | 2.40 |
ENSRNOT00000000629
|
Cpne5
|
copine 5 |
| chr1_-_215846911 | 2.29 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr8_-_59629133 | 1.90 |
ENSRNOT00000019458
|
Chrnb4
|
cholinergic receptor nicotinic beta 4 subunit |
| chr13_-_48284990 | 1.80 |
ENSRNOT00000086928
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr13_-_48284408 | 1.78 |
ENSRNOT00000085967
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr3_+_94530586 | 1.67 |
ENSRNOT00000067860
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr6_-_142993147 | 1.53 |
ENSRNOT00000044381
|
LOC100359978
|
mCG1038839-like |
| chr20_-_11528332 | 1.36 |
ENSRNOT00000038419
|
Tspear
|
thrombospondin-type laminin G domain and EAR repeats |
| chr20_+_48503973 | 1.35 |
ENSRNOT00000064081
|
Cdc40
|
cell division cycle 40 |
| chr10_+_14945265 | 1.16 |
ENSRNOT00000000246
|
Lmf1
|
lipase maturation factor 1 |
| chr10_-_87335823 | 1.12 |
ENSRNOT00000079297
|
Krt12
|
keratin 12 |
| chr18_+_24540659 | 0.89 |
ENSRNOT00000061074
|
Ammecr1l
|
AMMECR1 like |
| chr8_+_36909433 | 0.77 |
ENSRNOT00000074169
|
LOC100359924
|
prostate and testis expressed N-like |
| chr10_-_108425206 | 0.72 |
ENSRNOT00000073140
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chrX_-_17486721 | 0.51 |
ENSRNOT00000014611
|
Smptb
|
polypyrimidine tract-binding protein |
| chr10_+_84979717 | 0.49 |
ENSRNOT00000065555
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr12_-_35906382 | 0.45 |
ENSRNOT00000072179
|
Rack1
|
receptor for activated C kinase 1 |
| chr18_+_44468784 | 0.31 |
ENSRNOT00000031812
|
Dmxl1
|
Dmx-like 1 |
| chr9_-_113331319 | 0.27 |
ENSRNOT00000020681
|
Vapa
|
VAMP associated protein A |
| chr3_-_98092131 | 0.22 |
ENSRNOT00000006512
|
Fshb
|
follicle stimulating hormone beta subunit |
| chr4_+_117215064 | 0.17 |
ENSRNOT00000020909
|
Smyd5
|
SMYD family member 5 |
| chr1_+_174702373 | 0.13 |
ENSRNOT00000013733
|
Zfp143
|
zinc finger protein 143 |
| chr11_+_34598492 | 0.12 |
ENSRNOT00000065600
|
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr1_+_218058405 | 0.12 |
ENSRNOT00000028365
|
Fgf19
|
fibroblast growth factor 19 |
| chr17_+_42168263 | 0.12 |
ENSRNOT00000047422
|
LOC100360654
|
ribosomal protein L37-like |
| chr1_-_179010257 | 0.11 |
ENSRNOT00000017199
|
Rras2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr4_-_120390164 | 0.06 |
ENSRNOT00000017323
ENSRNOT00000063950 |
Eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr10_+_64336200 | 0.06 |
ENSRNOT00000046519
|
Rpl37
|
ribosomal protein L37 |
| chr11_-_17340373 | 0.05 |
ENSRNOT00000089762
|
AABR07033324.1
|
|
| chr5_+_18963937 | 0.05 |
ENSRNOT00000082188
|
AABR07047089.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Egr3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.9 | GO:0046864 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 2.4 | 17.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 1.8 | 5.4 | GO:1902996 | regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.2 | 3.6 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.7 | 4.3 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.7 | 3.4 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.6 | 1.9 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.3 | 2.7 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.3 | 2.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 0.7 | GO:0072714 | response to selenite ion(GO:0072714) regulation of selenocysteine incorporation(GO:1904569) |
| 0.2 | 1.2 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.2 | 7.6 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.4 | GO:0033123 | positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 1.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.3 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.4 | 2.7 | GO:0060205 | cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.3 | 3.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 5.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.4 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.1 | 1.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.0 | 12.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 5.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 4.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 5.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.8 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 2.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 5.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 3.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 5.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 3.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 17.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 2.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 1.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 3.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 5.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |