Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
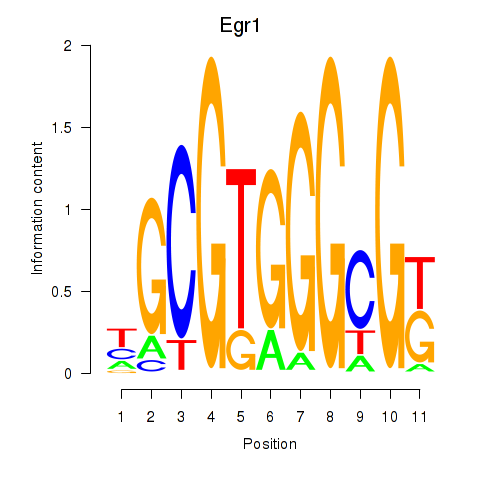
Results for Egr1
Z-value: 1.41
Transcription factors associated with Egr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr1
|
ENSRNOG00000019422 | early growth response 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr1 | rn6_v1_chr18_+_27657628_27657628 | 0.37 | 6.4e-12 | Click! |
Activity profile of Egr1 motif
Sorted Z-values of Egr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_22211071 | 69.15 |
ENSRNOT00000087817
ENSRNOT00000001910 |
Actl6b
|
actin-like 6B |
| chr14_-_86297623 | 57.92 |
ENSRNOT00000067162
ENSRNOT00000081607 ENSRNOT00000085265 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II beta |
| chr11_+_86512797 | 55.44 |
ENSRNOT00000051680
|
Gp1bb
|
glycoprotein Ib platelet beta subunit |
| chr8_+_22050222 | 53.53 |
ENSRNOT00000028096
|
Icam5
|
intercellular adhesion molecule 5 |
| chr1_-_103256823 | 48.86 |
ENSRNOT00000018860
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr3_+_155103150 | 48.78 |
ENSRNOT00000020720
|
Slc32a1
|
solute carrier family 32 member 1 |
| chr1_+_144070754 | 48.13 |
ENSRNOT00000079989
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr12_-_22680630 | 46.49 |
ENSRNOT00000041808
|
Vgf
|
VGF nerve growth factor inducible |
| chr7_-_119768082 | 46.15 |
ENSRNOT00000009612
|
Sstr3
|
somatostatin receptor 3 |
| chrX_+_157150655 | 45.21 |
ENSRNOT00000090795
|
Pnck
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr1_-_101360971 | 42.72 |
ENSRNOT00000028164
|
Lin7b
|
lin-7 homolog B, crumbs cell polarity complex component |
| chr8_-_40023193 | 42.63 |
ENSRNOT00000014404
|
Nrgn
|
neurogranin |
| chrX_+_122808605 | 42.03 |
ENSRNOT00000017567
|
Zcchc12
|
zinc finger CCHC-type containing 12 |
| chr1_+_165237847 | 41.31 |
ENSRNOT00000022963
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr6_+_132246602 | 40.60 |
ENSRNOT00000009896
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chrX_+_1321315 | 40.43 |
ENSRNOT00000014250
|
Syn1
|
synapsin I |
| chr3_-_2534663 | 37.68 |
ENSRNOT00000049297
ENSRNOT00000044246 |
Grin1
|
glutamate ionotropic receptor NMDA type subunit 1 |
| chr4_-_90882285 | 36.82 |
ENSRNOT00000039247
|
Snca
|
synuclein alpha |
| chr2_-_188645196 | 36.79 |
ENSRNOT00000083793
|
Efna3
|
ephrin A3 |
| chr7_-_59514939 | 36.13 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr3_-_66417741 | 36.05 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr10_-_85517683 | 35.64 |
ENSRNOT00000016070
|
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr8_-_120446455 | 35.30 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr7_-_105592804 | 35.06 |
ENSRNOT00000006789
|
Adcy8
|
adenylate cyclase 8 |
| chr13_-_15395617 | 34.21 |
ENSRNOT00000046060
|
LOC304725
|
similar to contactin associated protein-like 5 isoform 1 |
| chr10_+_86303727 | 34.07 |
ENSRNOT00000037752
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr7_+_122456799 | 33.91 |
ENSRNOT00000025564
|
Mchr1
|
melanin-concentrating hormone receptor 1 |
| chr9_+_73493027 | 33.84 |
ENSRNOT00000074427
ENSRNOT00000089478 |
Unc80
|
unc-80 homolog, NALCN activator |
| chr10_+_55940533 | 33.83 |
ENSRNOT00000012061
|
RGD1563441
|
similar to RIKEN cDNA A030009H04 |
| chr3_-_1924827 | 33.33 |
ENSRNOT00000006162
|
Cacna1b
|
calcium voltage-gated channel subunit alpha1 B |
| chr8_-_97115027 | 32.98 |
ENSRNOT00000018685
|
Ankrd34c
|
ankyrin repeat domain 34C |
| chr1_-_72727112 | 32.93 |
ENSRNOT00000031172
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr3_+_177188044 | 32.88 |
ENSRNOT00000022073
|
Tcea2
|
transcription elongation factor A2 |
| chr3_-_176644951 | 32.85 |
ENSRNOT00000049961
|
Kcnq2
|
potassium voltage-gated channel subfamily Q member 2 |
| chr12_-_22478752 | 32.83 |
ENSRNOT00000089392
ENSRNOT00000086915 |
Ache
|
acetylcholinesterase |
| chr7_+_73588163 | 32.28 |
ENSRNOT00000015707
|
Kcns2
|
potassium voltage-gated channel, modifier subfamily S, member 2 |
| chr3_+_154395187 | 31.97 |
ENSRNOT00000050810
|
Vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr1_-_64350338 | 31.70 |
ENSRNOT00000078444
|
Cacng8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chr1_+_263186235 | 28.34 |
ENSRNOT00000021876
|
Cnnm1
|
cyclin and CBS domain divalent metal cation transport mediator 1 |
| chr8_-_110813000 | 27.42 |
ENSRNOT00000010634
|
Ephb1
|
Eph receptor B1 |
| chr2_+_113984824 | 27.13 |
ENSRNOT00000086399
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chrX_+_39711201 | 26.69 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr7_+_78092037 | 25.98 |
ENSRNOT00000050753
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr2_+_113984646 | 25.04 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr14_+_60123169 | 25.04 |
ENSRNOT00000006610
|
Sel1l3
|
SEL1L family member 3 |
| chr7_-_140437467 | 24.87 |
ENSRNOT00000087181
|
Fkbp11
|
FK506 binding protein 11 |
| chr1_-_94494980 | 24.69 |
ENSRNOT00000020014
|
Ccne1
|
cyclin E1 |
| chr7_-_115910522 | 23.99 |
ENSRNOT00000076998
ENSRNOT00000067442 |
Arc
|
activity-regulated cytoskeleton-associated protein |
| chr5_-_144779212 | 23.98 |
ENSRNOT00000016230
|
Ncdn
|
neurochondrin |
| chr11_+_83975367 | 23.91 |
ENSRNOT00000058131
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr1_-_78440738 | 23.80 |
ENSRNOT00000020855
|
Npas1
|
neuronal PAS domain protein 1 |
| chr1_-_198454914 | 23.17 |
ENSRNOT00000049044
|
Prrt2
|
proline-rich transmembrane protein 2 |
| chrX_-_15467875 | 23.09 |
ENSRNOT00000011207
|
Pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr10_-_65766050 | 23.04 |
ENSRNOT00000013639
|
Sarm1
|
sterile alpha and TIR motif containing 1 |
| chr1_-_64405149 | 22.18 |
ENSRNOT00000089944
|
Cacng7
|
calcium voltage-gated channel auxiliary subunit gamma 7 |
| chr10_+_56627411 | 22.05 |
ENSRNOT00000089108
ENSRNOT00000068493 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr2_+_34186091 | 21.47 |
ENSRNOT00000016129
|
Sgtb
|
small glutamine rich tetratricopeptide repeat containing beta |
| chr2_+_198721724 | 21.41 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr2_-_26699333 | 21.35 |
ENSRNOT00000024459
|
Sv2c
|
synaptic vesicle glycoprotein 2c |
| chr2_-_186245771 | 20.02 |
ENSRNOT00000066004
ENSRNOT00000079954 |
Dclk2
|
doublecortin-like kinase 2 |
| chr7_-_84023316 | 19.49 |
ENSRNOT00000005556
|
Kcnv1
|
potassium voltage-gated channel modifier subfamily V member 1 |
| chr1_-_220467159 | 19.04 |
ENSRNOT00000075365
|
Tmem151a
|
transmembrane protein 151A |
| chr10_-_88667345 | 18.68 |
ENSRNOT00000025518
|
Kcnh4
|
potassium voltage-gated channel subfamily H member 4 |
| chr2_-_186245342 | 18.67 |
ENSRNOT00000057062
ENSRNOT00000022292 |
Dclk2
|
doublecortin-like kinase 2 |
| chr4_-_153028129 | 18.55 |
ENSRNOT00000074558
|
Cecr6
|
cat eye syndrome chromosome region, candidate 6 |
| chr5_-_63192025 | 18.16 |
ENSRNOT00000008913
|
Alg2
|
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chr4_+_154309426 | 18.06 |
ENSRNOT00000019346
|
A2m
|
alpha-2-macroglobulin |
| chr8_+_65611570 | 17.86 |
ENSRNOT00000017147
|
Larp6
|
La ribonucleoprotein domain family, member 6 |
| chr1_-_143278485 | 17.58 |
ENSRNOT00000026009
|
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr2_-_184263564 | 17.20 |
ENSRNOT00000015279
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr6_-_1319541 | 16.23 |
ENSRNOT00000006527
|
Strn
|
striatin |
| chr2_-_147959567 | 15.50 |
ENSRNOT00000063986
|
LOC100909840
|
profilin-2-like |
| chr16_-_20860767 | 15.36 |
ENSRNOT00000027275
ENSRNOT00000092417 |
Gdf1
Cers1
|
growth differentiation factor 1 ceramide synthase 1 |
| chr16_-_22350155 | 15.30 |
ENSRNOT00000015931
|
Atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr3_+_146546387 | 15.30 |
ENSRNOT00000009946
|
Entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 |
| chr6_+_135856218 | 14.64 |
ENSRNOT00000066715
|
RGD1560608
|
similar to novel protein |
| chr5_+_1417478 | 14.48 |
ENSRNOT00000008153
ENSRNOT00000085564 |
Jph1
|
junctophilin 1 |
| chrX_+_136466779 | 14.45 |
ENSRNOT00000093268
ENSRNOT00000068717 |
Arhgap36
|
Rho GTPase activating protein 36 |
| chr14_-_11198194 | 14.34 |
ENSRNOT00000003083
|
Enoph1
|
enolase-phosphatase 1 |
| chr1_-_162300515 | 14.32 |
ENSRNOT00000031161
|
Usp35
|
ubiquitin specific peptidase 35 |
| chr2_+_116416507 | 14.17 |
ENSRNOT00000030700
|
Actrt3
|
actin-related protein T3 |
| chr5_+_162031722 | 13.92 |
ENSRNOT00000020483
|
Lrrc38
|
leucine rich repeat containing 38 |
| chr3_-_114307250 | 13.84 |
ENSRNOT00000064592
|
Shf
|
Src homology 2 domain containing F |
| chr4_+_71675383 | 13.76 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr7_+_11201690 | 13.39 |
ENSRNOT00000005624
|
Fzr1
|
fizzy/cell division cycle 20 related 1 |
| chr9_+_81783349 | 13.28 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr4_+_154423209 | 12.55 |
ENSRNOT00000075799
|
LOC100911545
|
alpha-2-macroglobulin-like |
| chr5_-_166726794 | 12.37 |
ENSRNOT00000022799
|
Slc25a33
|
solute carrier family 25 member 33 |
| chr16_-_9709347 | 12.12 |
ENSRNOT00000083933
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr3_-_111560556 | 12.06 |
ENSRNOT00000030532
|
Ltk
|
leukocyte receptor tyrosine kinase |
| chr10_+_55675575 | 12.04 |
ENSRNOT00000057295
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr1_-_142164007 | 11.48 |
ENSRNOT00000078982
|
Man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chrX_-_30831483 | 11.37 |
ENSRNOT00000004443
|
Glra2
|
glycine receptor, alpha 2 |
| chr3_-_92290919 | 11.13 |
ENSRNOT00000007077
|
Fjx1
|
four jointed box 1 |
| chr9_-_44456554 | 11.07 |
ENSRNOT00000080011
|
Tsga10
|
testis specific 10 |
| chr15_-_61772516 | 10.93 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr10_+_13836128 | 10.92 |
ENSRNOT00000012720
|
Pgp
|
phosphoglycolate phosphatase |
| chr9_+_82033543 | 10.47 |
ENSRNOT00000023439
|
Wnt6
|
wingless-type MMTV integration site family, member 6 |
| chr3_-_171134655 | 10.19 |
ENSRNOT00000028960
|
RGD1561252
|
similar to Ubiquitin carboxyl-terminal hydrolase isozyme L3 (Ubiquitin thiolesterase L3) |
| chr1_-_142164263 | 9.68 |
ENSRNOT00000016281
|
Man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr5_-_101588598 | 9.46 |
ENSRNOT00000082239
|
Psip1
|
PC4 and SFRS1 interacting protein 1 |
| chr5_-_142845116 | 9.46 |
ENSRNOT00000065105
|
RGD1559909
|
RGD1559909 |
| chr7_-_144109116 | 9.32 |
ENSRNOT00000020437
|
Map3k12
|
mitogen activated protein kinase kinase kinase 12 |
| chr4_-_16130563 | 9.26 |
ENSRNOT00000090240
ENSRNOT00000034969 |
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr12_-_46920952 | 9.20 |
ENSRNOT00000001532
|
Msi1
|
musashi RNA-binding protein 1 |
| chr1_+_220335254 | 9.13 |
ENSRNOT00000072261
|
Rin1
|
Ras and Rab interactor 1 |
| chrX_+_17540458 | 9.01 |
ENSRNOT00000045710
|
Nudt11
|
nudix hydrolase 11 |
| chr4_+_56744561 | 8.82 |
ENSRNOT00000009737
|
Atp6v1f
|
ATPase H+ transporting V1 subunit F |
| chr10_+_109107389 | 8.73 |
ENSRNOT00000068437
ENSRNOT00000005687 |
Baiap2
|
BAI1-associated protein 2 |
| chr11_+_69484293 | 8.68 |
ENSRNOT00000049292
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr8_+_63379087 | 8.51 |
ENSRNOT00000012091
ENSRNOT00000012295 |
Nptn
|
neuroplastin |
| chr9_-_119818310 | 8.45 |
ENSRNOT00000019359
|
Smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr6_+_27975849 | 8.34 |
ENSRNOT00000060810
|
Dtnb
|
dystrobrevin, beta |
| chr1_-_198128857 | 8.28 |
ENSRNOT00000026496
|
Coro1a
|
coronin 1A |
| chr3_+_151553578 | 8.16 |
ENSRNOT00000045484
|
Ergic3
|
ERGIC and golgi 3 |
| chr15_+_33121273 | 8.15 |
ENSRNOT00000016020
|
Rem2
|
RRAD and GEM like GTPase 2 |
| chr1_+_220428481 | 7.98 |
ENSRNOT00000027335
|
LOC108348044
|
ras and Rab interactor 1 |
| chr12_+_2589818 | 7.95 |
ENSRNOT00000087848
|
AABR07034980.1
|
|
| chr5_+_147246037 | 7.89 |
ENSRNOT00000089457
|
Rnf19b
|
ring finger protein 19B |
| chr4_+_117215064 | 7.80 |
ENSRNOT00000020909
|
Smyd5
|
SMYD family member 5 |
| chr15_+_86153628 | 7.73 |
ENSRNOT00000012842
|
Uchl3
|
ubiquitin C-terminal hydrolase L3 |
| chrX_-_156379189 | 7.61 |
ENSRNOT00000083148
|
Plxna3
|
plexin A3 |
| chr12_+_7865938 | 7.52 |
ENSRNOT00000061229
|
Ubl3
|
ubiquitin-like 3 |
| chr10_-_83128297 | 7.41 |
ENSRNOT00000082160
|
Kat7
|
lysine acetyltransferase 7 |
| chr6_-_99783047 | 7.35 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr3_-_110492398 | 7.31 |
ENSRNOT00000056450
|
Ankrd63
|
ankyrin repeat domain 63 |
| chr4_-_16130848 | 7.21 |
ENSRNOT00000042914
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr5_-_99033107 | 7.16 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr8_+_116154736 | 7.10 |
ENSRNOT00000021218
|
Cacna2d2
|
calcium voltage-gated channel auxiliary subunit alpha2delta 2 |
| chr6_+_27975417 | 7.10 |
ENSRNOT00000077830
|
Dtnb
|
dystrobrevin, beta |
| chr1_-_215836641 | 6.95 |
ENSRNOT00000080246
|
Igf2
|
insulin-like growth factor 2 |
| chr1_+_193335645 | 6.80 |
ENSRNOT00000019640
|
Lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr7_+_41115154 | 6.76 |
ENSRNOT00000066174
|
Atp2b1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr6_-_6842758 | 6.73 |
ENSRNOT00000006094
|
Kcng3
|
potassium voltage-gated channel modifier subfamily G member 3 |
| chr11_-_17340373 | 6.68 |
ENSRNOT00000089762
|
AABR07033324.1
|
|
| chr8_+_128577345 | 6.57 |
ENSRNOT00000082356
|
Wdr48
|
WD repeat domain 48 |
| chr1_+_98440186 | 6.31 |
ENSRNOT00000024150
|
Iglon5
|
IgLON family member 5 |
| chr20_+_32509598 | 6.27 |
ENSRNOT00000046268
|
Kpna5
|
karyopherin subunit alpha 5 |
| chr10_-_105552986 | 6.17 |
ENSRNOT00000014699
|
Ube2o
|
ubiquitin-conjugating enzyme E2O |
| chr16_-_61753476 | 6.13 |
ENSRNOT00000016792
|
Saraf
|
store-operated calcium entry-associated regulatory factor |
| chr1_+_64031859 | 5.91 |
ENSRNOT00000090873
|
Mboat7l1
|
membrane bound O-acyltransferase domain containing 7-like 1 |
| chr8_+_44990014 | 5.78 |
ENSRNOT00000044608
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr7_+_12006710 | 5.63 |
ENSRNOT00000045421
|
Klf16
|
Kruppel-like factor 16 |
| chr7_+_11545024 | 5.55 |
ENSRNOT00000073221
|
Slc39a3
|
solute carrier family 39 member 3 |
| chr8_+_128577080 | 5.32 |
ENSRNOT00000024172
|
Wdr48
|
WD repeat domain 48 |
| chr15_+_27850151 | 5.25 |
ENSRNOT00000087958
|
Apex1
|
apurinic/apyrimidinic endodeoxyribonuclease 1 |
| chr7_-_94755924 | 5.16 |
ENSRNOT00000040836
|
Taf2
|
TATA-box binding protein associated factor 2 |
| chr10_+_63731959 | 4.93 |
ENSRNOT00000090554
|
Pitpna
|
phosphatidylinositol transfer protein, alpha |
| chr9_-_11240282 | 4.93 |
ENSRNOT00000070920
|
Uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr3_+_71020534 | 4.87 |
ENSRNOT00000007276
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr8_-_82351108 | 4.86 |
ENSRNOT00000013053
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr10_-_13515448 | 4.79 |
ENSRNOT00000067660
|
Pdpk1
|
3-phosphoinositide dependent protein kinase-1 |
| chr4_-_87111025 | 4.75 |
ENSRNOT00000018442
|
Kbtbd2
|
kelch repeat and BTB domain containing 2 |
| chr3_-_163935617 | 4.73 |
ENSRNOT00000074023
|
Kcnb1
|
potassium voltage-gated channel subfamily B member 1 |
| chr1_-_220203532 | 4.72 |
ENSRNOT00000079984
|
Peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr3_+_8534440 | 4.57 |
ENSRNOT00000045827
ENSRNOT00000082672 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr9_+_47386626 | 4.48 |
ENSRNOT00000021270
|
Slc9a2
|
solute carrier family 9 member A2 |
| chr18_+_27554062 | 4.25 |
ENSRNOT00000088432
|
LOC100363294
|
stem-loop binding protein-like |
| chr1_-_82108083 | 4.25 |
ENSRNOT00000027677
ENSRNOT00000084530 |
Gsk3a
|
glycogen synthase kinase 3 alpha |
| chr10_-_47857326 | 4.05 |
ENSRNOT00000085703
ENSRNOT00000091963 |
Epn2
|
epsin 2 |
| chr3_+_176093640 | 4.04 |
ENSRNOT00000086541
|
Ogfr
|
opioid growth factor receptor |
| chr20_+_13940877 | 3.92 |
ENSRNOT00000093587
|
Cabin1
|
calcineurin binding protein 1 |
| chr16_+_27399467 | 3.89 |
ENSRNOT00000065642
|
Tll1
|
tolloid-like 1 |
| chr6_-_80334522 | 3.85 |
ENSRNOT00000059316
|
Fbxo33
|
F-box protein 33 |
| chr1_-_36319896 | 3.82 |
ENSRNOT00000023641
ENSRNOT00000090884 |
Nsun2
|
NOP2/Sun RNA methyltransferase family, member 2 |
| chr16_+_53957858 | 3.74 |
ENSRNOT00000013078
|
Frg1
|
FSHD region gene 1 |
| chr14_+_11198896 | 3.68 |
ENSRNOT00000079767
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr7_-_117722703 | 3.68 |
ENSRNOT00000093083
ENSRNOT00000092789 |
Cyhr1
|
cysteine and histidine rich 1 |
| chr10_-_104263071 | 3.43 |
ENSRNOT00000005347
|
Grb2
|
growth factor receptor bound protein 2 |
| chr8_-_36467627 | 3.33 |
ENSRNOT00000082346
|
Fam118b
|
family with sequence similarity 118, member B |
| chr8_-_48762342 | 3.33 |
ENSRNOT00000049125
|
Foxr1
|
forkhead box R1 |
| chr1_-_226353611 | 3.24 |
ENSRNOT00000037624
|
Dagla
|
diacylglycerol lipase, alpha |
| chr1_-_219422268 | 3.13 |
ENSRNOT00000025092
|
Carns1
|
carnosine synthase 1 |
| chr5_+_58393233 | 3.07 |
ENSRNOT00000000142
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr8_+_75516904 | 3.05 |
ENSRNOT00000013142
|
Rora
|
RAR-related orphan receptor A |
| chr5_+_58393603 | 2.99 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chrX_-_14890606 | 2.99 |
ENSRNOT00000049864
|
RGD1560784
|
similar to RIKEN cDNA B630019K06 |
| chr13_+_51958834 | 2.94 |
ENSRNOT00000007833
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr13_+_74154835 | 2.91 |
ENSRNOT00000059524
|
Abl2
|
ABL proto-oncogene 2, non-receptor tyrosine kinase |
| chr7_+_123381077 | 2.91 |
ENSRNOT00000082603
ENSRNOT00000056041 |
Srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr3_+_175930505 | 2.89 |
ENSRNOT00000012611
|
Ogfr
|
opioid growth factor receptor |
| chr17_+_6785428 | 2.84 |
ENSRNOT00000026070
|
Gkap1
|
G kinase anchoring protein 1 |
| chr12_+_22726643 | 2.82 |
ENSRNOT00000040015
|
Znhit1
|
zinc finger, HIT-type containing 1 |
| chr1_-_114371897 | 2.80 |
ENSRNOT00000018028
|
Nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 |
| chrX_+_120859968 | 2.69 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr2_+_209433103 | 2.69 |
ENSRNOT00000024036
|
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr1_+_116587815 | 2.55 |
ENSRNOT00000021366
|
Ube3a
|
ubiquitin protein ligase E3A |
| chr1_+_220009397 | 2.44 |
ENSRNOT00000084410
|
Rbm4b
|
RNA binding motif protein 4B |
| chr10_+_61685241 | 2.41 |
ENSRNOT00000092606
|
Mnt
|
MAX network transcriptional repressor |
| chr12_-_9331195 | 2.05 |
ENSRNOT00000044134
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chrX_+_120860178 | 2.02 |
ENSRNOT00000088661
|
Wdr44
|
WD repeat domain 44 |
| chr3_+_62648447 | 1.89 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chr11_+_34101197 | 1.83 |
ENSRNOT00000002299
|
Chaf1b
|
chromatin assembly factor 1 subunit B |
| chr13_+_56015901 | 1.74 |
ENSRNOT00000043907
|
Dennd1b
|
DENN domain containing 1B |
| chr18_+_36596585 | 1.65 |
ENSRNOT00000036613
|
Rbm27
|
RNA binding motif protein 27 |
| chr5_-_136112344 | 1.63 |
ENSRNOT00000050195
|
RGD1563714
|
RGD1563714 |
| chr6_-_26497328 | 1.62 |
ENSRNOT00000074938
|
Nrbp1
|
nuclear receptor binding protein 1 |
| chr14_-_17436897 | 1.57 |
ENSRNOT00000003277
|
Uso1
|
USO1 vesicle transport factor |
| chr8_-_48564722 | 1.51 |
ENSRNOT00000067902
|
Cbl
|
Cbl proto-oncogene |
| chr3_-_15433252 | 1.37 |
ENSRNOT00000008817
|
Lhx6
|
LIM homeobox 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Egr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.3 | 57.9 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 15.4 | 15.4 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 12.6 | 37.7 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 12.3 | 36.8 | GO:2000468 | regulation of hydrogen peroxide catabolic process(GO:2000295) regulation of peroxidase activity(GO:2000468) |
| 11.1 | 55.3 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 8.1 | 48.9 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 7.7 | 46.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 7.6 | 61.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 7.2 | 36.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 7.1 | 42.7 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 5.7 | 17.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 5.4 | 48.8 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 4.9 | 34.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 4.4 | 22.0 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 4.4 | 8.7 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 4.3 | 8.7 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 4.0 | 40.4 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 4.0 | 12.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 3.7 | 52.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 3.7 | 7.4 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 3.6 | 32.8 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 3.6 | 18.1 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 3.2 | 3.2 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 3.1 | 40.6 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 2.8 | 8.5 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 2.8 | 8.5 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 2.7 | 16.5 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 2.7 | 10.9 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 2.7 | 70.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 2.7 | 18.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 2.4 | 46.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 2.4 | 14.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 2.4 | 11.9 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 2.3 | 27.4 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 2.3 | 43.4 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 2.3 | 9.0 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 2.1 | 4.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 2.1 | 44.3 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 2.1 | 10.5 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 2.1 | 33.3 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 2.1 | 24.7 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 2.0 | 36.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 2.0 | 14.0 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 2.0 | 26.0 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 2.0 | 37.5 | GO:0043084 | penile erection(GO:0043084) |
| 2.0 | 17.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.9 | 23.0 | GO:0042711 | maternal behavior(GO:0042711) |
| 1.8 | 9.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 1.8 | 10.9 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 1.8 | 18.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 1.7 | 6.8 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 1.6 | 4.7 | GO:0045751 | regulation of Toll signaling pathway(GO:0008592) negative regulation of Toll signaling pathway(GO:0045751) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 1.5 | 12.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 1.4 | 7.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 1.4 | 8.3 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 1.3 | 15.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.3 | 11.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 1.2 | 37.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 1.2 | 23.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 1.2 | 9.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 1.1 | 3.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 1.1 | 13.8 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.0 | 17.0 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 1.0 | 7.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.0 | 2.9 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 1.0 | 32.7 | GO:0007616 | long-term memory(GO:0007616) |
| 0.8 | 10.9 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.8 | 15.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.6 | 7.7 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.6 | 15.3 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.6 | 24.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.5 | 32.9 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.5 | 10.8 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.5 | 4.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.5 | 6.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.5 | 11.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.4 | 3.8 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.4 | 51.1 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.4 | 33.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.4 | 8.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 23.1 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.3 | 4.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 22.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.3 | 6.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.3 | 0.6 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.3 | 7.7 | GO:0042755 | eating behavior(GO:0042755) |
| 0.3 | 1.1 | GO:0015746 | citrate transport(GO:0015746) |
| 0.3 | 5.2 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.3 | 1.4 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 | 4.9 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.3 | 23.0 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.3 | 1.9 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 4.9 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.3 | 23.9 | GO:0048167 | regulation of synaptic plasticity(GO:0048167) |
| 0.3 | 4.2 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.3 | 1.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 3.1 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.3 | 7.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.2 | 11.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 22.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.2 | 1.2 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.2 | 32.0 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.2 | 2.5 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.2 | 0.4 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 | 5.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 2.0 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 3.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.2 | 1.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 6.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 1.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.2 | 5.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 5.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.4 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 1.7 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 13.7 | GO:0016358 | dendrite development(GO:0016358) |
| 0.1 | 21.5 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 4.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 4.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 11.6 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.1 | 3.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) selenocysteine metabolic process(GO:0016259) |
| 0.1 | 1.0 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.1 | 0.2 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 6.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.4 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 1.7 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 2.1 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.2 | GO:0010829 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.4 | GO:0060976 | coronary vasculature development(GO:0060976) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.8 | 48.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 9.4 | 37.7 | GO:0044307 | dendritic branch(GO:0044307) |
| 9.2 | 36.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 4.8 | 14.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 4.8 | 66.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 4.6 | 23.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 4.4 | 48.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 4.3 | 12.9 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 3.4 | 17.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 3.3 | 36.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 2.7 | 32.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 2.7 | 13.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 2.4 | 22.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 2.4 | 12.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 2.0 | 24.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 1.9 | 53.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 1.8 | 57.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 1.5 | 27.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 1.4 | 35.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.4 | 66.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 1.3 | 11.9 | GO:0008091 | spectrin(GO:0008091) |
| 1.3 | 72.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 1.3 | 32.9 | GO:0043194 | node of Ranvier(GO:0033268) axon initial segment(GO:0043194) |
| 1.2 | 17.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.1 | 6.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 1.1 | 45.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 1.1 | 8.5 | GO:0001740 | Barr body(GO:0001740) |
| 1.0 | 7.9 | GO:0044194 | cytolytic granule(GO:0044194) |
| 1.0 | 2.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.9 | 3.4 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.8 | 81.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.8 | 164.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.8 | 17.3 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.7 | 13.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.6 | 48.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.6 | 14.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.6 | 1.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.6 | 11.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.6 | 46.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.5 | 12.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.5 | 12.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.4 | 9.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.4 | 4.9 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.4 | 5.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.4 | 21.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 4.1 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.2 | 0.7 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 0.4 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.2 | 3.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 9.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 2.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 8.7 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 13.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 6.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 34.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 11.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 11.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 4.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 5.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 24.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 3.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 25.9 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 2.9 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 6.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 15.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 2.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.4 | 34.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 10.9 | 32.8 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 9.8 | 48.8 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 7.7 | 46.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 7.5 | 37.7 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 6.1 | 18.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 6.0 | 23.9 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 5.8 | 35.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 5.5 | 16.5 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 4.5 | 18.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 4.3 | 99.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 3.8 | 15.3 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 3.8 | 11.4 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 3.7 | 22.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 3.4 | 41.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 3.4 | 17.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 3.2 | 12.9 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 2.8 | 42.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 2.8 | 36.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 2.7 | 13.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 2.7 | 18.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 2.4 | 33.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 2.3 | 27.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 2.3 | 9.0 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 1.9 | 15.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 1.8 | 42.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 1.8 | 10.9 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 1.7 | 5.2 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 1.7 | 8.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.7 | 8.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.6 | 83.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 1.6 | 16.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 1.5 | 15.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.5 | 46.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 1.5 | 12.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 1.4 | 45.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 1.4 | 17.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 1.2 | 36.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 1.2 | 9.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 1.1 | 48.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 1.0 | 40.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 1.0 | 5.2 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 1.0 | 3.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.0 | 4.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.0 | 6.8 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 1.0 | 3.8 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.8 | 36.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.8 | 78.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.8 | 30.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.8 | 8.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.7 | 52.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.7 | 13.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.6 | 20.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.6 | 24.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.5 | 7.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.5 | 7.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 4.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 10.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.4 | 57.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.4 | 53.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.4 | 8.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.4 | 8.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.4 | 7.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.4 | 14.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.4 | 3.9 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.4 | 3.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.4 | 6.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 4.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.3 | 1.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 0.3 | 10.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 4.6 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.2 | 6.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 5.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 14.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 2.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 3.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 16.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 4.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 32.6 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 1.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.4 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.1 | 0.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 5.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 57.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.7 | 35.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 1.5 | 24.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.2 | 34.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 1.0 | 27.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 1.0 | 36.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.9 | 86.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.9 | 17.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.9 | 8.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.7 | 28.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.6 | 15.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.6 | 12.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.6 | 18.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.6 | 9.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.5 | 28.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.5 | 4.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 16.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.5 | 2.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 9.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 4.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.4 | 16.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 5.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 4.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 11.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 8.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 0.7 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.2 | 1.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 10.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 7.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 96.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 4.1 | 61.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 3.7 | 55.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 3.5 | 52.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 3.4 | 40.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 2.3 | 35.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 1.9 | 9.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 1.9 | 96.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 1.4 | 36.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 1.3 | 36.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 1.3 | 32.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 1.3 | 18.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.2 | 34.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 1.1 | 24.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.0 | 31.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.9 | 24.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.9 | 36.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.8 | 14.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.8 | 12.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.7 | 15.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.6 | 18.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.5 | 7.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.5 | 17.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.5 | 4.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.5 | 4.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.5 | 4.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.5 | 80.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.4 | 4.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.4 | 11.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.4 | 5.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.4 | 4.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 7.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 31.5 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.3 | 5.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 5.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 7.4 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.1 | 10.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 3.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 7.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 6.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 4.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 3.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |