Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Ebf3
Z-value: 1.33
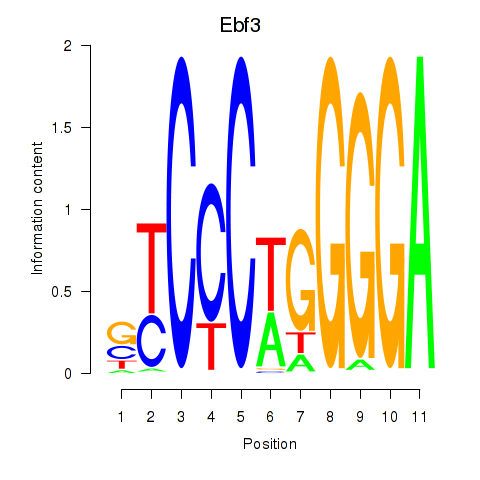
Transcription factors associated with Ebf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ebf3
|
ENSRNOG00000016102 | early B-cell factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ebf3 | rn6_v1_chr1_-_209641123_209641123 | 0.28 | 4.5e-07 | Click! |
Activity profile of Ebf3 motif
Sorted Z-values of Ebf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_98371184 | 55.10 |
ENSRNOT00000086911
|
AABR07060872.1
|
|
| chr13_+_52667969 | 47.63 |
ENSRNOT00000084986
ENSRNOT00000050284 |
Tnnt2
|
troponin T2, cardiac type |
| chr2_+_205160405 | 45.25 |
ENSRNOT00000035605
|
Tspan2
|
tetraspanin 2 |
| chr10_-_15603649 | 43.48 |
ENSRNOT00000051483
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr4_+_98370797 | 39.91 |
ENSRNOT00000031991
|
AABR07060872.1
|
|
| chr9_+_82596355 | 39.80 |
ENSRNOT00000065076
|
Speg
|
SPEG complex locus |
| chr6_-_138632159 | 39.42 |
ENSRNOT00000082921
ENSRNOT00000040702 |
Ighm
|
immunoglobulin heavy constant mu |
| chr1_+_80321585 | 37.32 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr9_-_19372673 | 33.09 |
ENSRNOT00000073667
ENSRNOT00000079517 |
Clic5
|
chloride intracellular channel 5 |
| chr3_+_19045214 | 33.08 |
ENSRNOT00000070878
|
AABR07051670.1
|
|
| chr4_+_101645731 | 31.78 |
ENSRNOT00000087901
|
AABR07060953.1
|
|
| chr4_+_102147211 | 29.46 |
ENSRNOT00000083239
|
AABR07060980.1
|
|
| chr6_+_139405966 | 28.17 |
ENSRNOT00000088974
|
AABR07065693.3
|
|
| chrX_+_159158194 | 27.03 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr1_+_81763614 | 24.79 |
ENSRNOT00000027254
|
Cd79a
|
CD79a molecule |
| chr6_-_138900915 | 24.38 |
ENSRNOT00000075363
|
AABR07065656.3
|
|
| chr7_-_119716238 | 23.39 |
ENSRNOT00000075678
|
Il2rb
|
interleukin 2 receptor subunit beta |
| chr6_-_138631997 | 23.37 |
ENSRNOT00000073304
|
AABR07065651.3
|
|
| chr1_-_82580007 | 22.34 |
ENSRNOT00000078668
|
Axl
|
Axl receptor tyrosine kinase |
| chr12_-_16126953 | 22.06 |
ENSRNOT00000001682
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_-_138685656 | 21.75 |
ENSRNOT00000041706
|
AABR07065651.7
|
|
| chr7_+_121841855 | 21.27 |
ENSRNOT00000024673
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr6_-_138736203 | 21.17 |
ENSRNOT00000052021
|
LOC100360169
|
rCG21044-like |
| chr3_+_19128400 | 20.99 |
ENSRNOT00000074272
|
AABR07051673.1
|
|
| chr4_-_103145058 | 20.97 |
ENSRNOT00000073076
|
AABR07061048.1
|
|
| chr4_+_102351036 | 20.46 |
ENSRNOT00000079277
|
AABR07060994.1
|
|
| chr9_+_47536824 | 20.44 |
ENSRNOT00000049349
|
Tmem182
|
transmembrane protein 182 |
| chr3_+_41019898 | 19.66 |
ENSRNOT00000007335
|
Kcnj3
|
potassium voltage-gated channel subfamily J member 3 |
| chr4_+_49369296 | 19.59 |
ENSRNOT00000007822
|
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr3_+_19141133 | 19.47 |
ENSRNOT00000058323
|
AABR07051675.1
|
|
| chrX_+_159112880 | 18.76 |
ENSRNOT00000084594
|
Fhl1
|
four and a half LIM domains 1 |
| chr3_+_19690016 | 18.60 |
ENSRNOT00000085460
|
AABR07051707.1
|
|
| chr6_-_140880070 | 18.45 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr3_+_19174027 | 18.39 |
ENSRNOT00000074445
|
AABR07051678.1
|
|
| chr14_+_84306466 | 18.11 |
ENSRNOT00000006116
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr13_-_91872954 | 18.07 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr20_+_29897594 | 18.03 |
ENSRNOT00000057752
|
Vsir
|
V-set immunoregulatory receptor |
| chrX_+_134979646 | 17.97 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr10_+_77537340 | 17.95 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chr4_+_102643934 | 17.77 |
ENSRNOT00000058389
|
LOC100911032
|
uncharacterized LOC100911032 |
| chr6_-_141198565 | 17.59 |
ENSRNOT00000064361
|
AABR07065782.1
|
|
| chr1_+_199495298 | 17.39 |
ENSRNOT00000086003
ENSRNOT00000026748 |
Itgad
|
integrin subunit alpha D |
| chr20_+_46199981 | 17.38 |
ENSRNOT00000000337
|
Mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr16_-_39719187 | 16.93 |
ENSRNOT00000092971
|
Gpm6a
|
glycoprotein m6a |
| chr6_-_140642221 | 16.80 |
ENSRNOT00000081996
|
AABR07065772.2
|
|
| chr20_+_4363508 | 16.61 |
ENSRNOT00000077205
|
Ager
|
advanced glycosylation end product-specific receptor |
| chr17_-_79676499 | 16.42 |
ENSRNOT00000022711
|
Itga8
|
integrin subunit alpha 8 |
| chr9_+_17340341 | 16.03 |
ENSRNOT00000026637
ENSRNOT00000026559 ENSRNOT00000042790 ENSRNOT00000044163 ENSRNOT00000083811 |
Vegfa
|
vascular endothelial growth factor A |
| chr6_-_139710905 | 16.01 |
ENSRNOT00000077430
|
AABR07065705.2
|
|
| chr18_-_6781841 | 15.74 |
ENSRNOT00000077606
ENSRNOT00000048109 |
Aqp4
|
aquaporin 4 |
| chr3_+_19366370 | 15.27 |
ENSRNOT00000086557
|
AABR07051689.1
|
|
| chr6_-_139004980 | 14.90 |
ENSRNOT00000085427
ENSRNOT00000087351 |
AABR07065656.4
|
|
| chr2_+_41442241 | 14.83 |
ENSRNOT00000067546
|
Pde4d
|
phosphodiesterase 4D |
| chr4_+_103495993 | 14.81 |
ENSRNOT00000072325
|
AABR07061068.1
|
|
| chr1_-_98521551 | 14.81 |
ENSRNOT00000081922
|
Siglec10
|
sialic acid binding Ig-like lectin 10 |
| chr6_+_139560028 | 14.60 |
ENSRNOT00000072633
|
LOC100360581
|
rCG58847-like |
| chr3_+_18787606 | 14.49 |
ENSRNOT00000090508
|
AABR07051658.1
|
|
| chr6_-_138954577 | 14.43 |
ENSRNOT00000042728
|
AABR07065656.7
|
|
| chr1_-_267245636 | 14.32 |
ENSRNOT00000082799
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr8_-_116361343 | 13.98 |
ENSRNOT00000066296
|
Sema3b
|
semaphorin 3B |
| chr6_-_139660666 | 13.81 |
ENSRNOT00000086098
|
AABR07065705.1
|
|
| chr6_-_138931429 | 13.64 |
ENSRNOT00000090584
|
LOC100359993
|
Ighg protein-like |
| chr8_-_63092009 | 13.53 |
ENSRNOT00000034300
|
Loxl1
|
lysyl oxidase-like 1 |
| chr5_+_156026911 | 13.50 |
ENSRNOT00000046802
|
Rap1gap
|
Rap1 GTPase-activating protein |
| chr6_-_142353308 | 13.45 |
ENSRNOT00000066416
|
AABR07065814.2
|
|
| chr13_+_85818427 | 13.44 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr2_+_187347602 | 13.41 |
ENSRNOT00000025384
|
Nes
|
nestin |
| chr1_+_264893162 | 13.30 |
ENSRNOT00000021714
|
Tlx1
|
T-cell leukemia, homeobox 1 |
| chr2_-_210550490 | 13.19 |
ENSRNOT00000081835
ENSRNOT00000025222 ENSRNOT00000086403 |
Csf1
|
colony stimulating factor 1 |
| chr5_-_141363524 | 13.17 |
ENSRNOT00000084109
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr4_-_122741110 | 12.79 |
ENSRNOT00000008888
|
Nup210
|
nucleoporin 210 |
| chr6_+_139293294 | 12.64 |
ENSRNOT00000050297
ENSRNOT00000081817 |
LOC100359993
|
Ighg protein-like |
| chr18_+_65388685 | 12.62 |
ENSRNOT00000080844
|
Tcf4
|
transcription factor 4 |
| chr16_-_20686317 | 12.56 |
ENSRNOT00000060097
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr7_+_143707237 | 12.49 |
ENSRNOT00000074212
|
Tns2
|
tensin 2 |
| chr18_+_17043903 | 12.46 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr4_-_159079003 | 12.28 |
ENSRNOT00000026691
|
Kcna5
|
potassium voltage-gated channel subfamily A member 5 |
| chr6_-_139637187 | 12.13 |
ENSRNOT00000089454
|
LOC100359993
|
Ighg protein-like |
| chr5_+_140692779 | 12.08 |
ENSRNOT00000019101
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr3_-_7795758 | 11.37 |
ENSRNOT00000045919
|
Ntng2
|
netrin G2 |
| chr10_+_92288910 | 10.92 |
ENSRNOT00000006947
ENSRNOT00000045127 |
Mapt
|
microtubule-associated protein tau |
| chr18_-_77322690 | 10.76 |
ENSRNOT00000058382
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr2_+_54191538 | 10.64 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr8_-_48619592 | 10.45 |
ENSRNOT00000012534
|
Abcg4
|
ATP binding cassette subfamily G member 4 |
| chr17_-_67945037 | 10.45 |
ENSRNOT00000023033
|
Klf6
|
Kruppel-like factor 6 |
| chr15_+_52148379 | 10.44 |
ENSRNOT00000074912
|
Phyhip
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chrX_+_15273933 | 10.38 |
ENSRNOT00000075082
|
LOC108348091
|
erythroid transcription factor |
| chr15_+_52767442 | 10.25 |
ENSRNOT00000014441
|
LOC108348161
|
phytanoyl-CoA hydroxylase-interacting protein |
| chr14_+_77712240 | 10.24 |
ENSRNOT00000009101
|
Msx1
|
msh homeobox 1 |
| chr12_-_21760292 | 10.22 |
ENSRNOT00000059592
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr10_+_92289107 | 9.95 |
ENSRNOT00000050070
|
Mapt
|
microtubule-associated protein tau |
| chr3_+_148234193 | 9.94 |
ENSRNOT00000010418
|
Cox4i2
|
cytochrome c oxidase subunit 4i2 |
| chr1_+_81230612 | 9.91 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr9_+_81656116 | 9.81 |
ENSRNOT00000083421
|
Slc11a1
|
solute carrier family 11 member 1 |
| chr5_-_173611202 | 9.76 |
ENSRNOT00000047854
|
Agrn
|
agrin |
| chr10_-_83655182 | 9.71 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr6_-_142060032 | 9.65 |
ENSRNOT00000064717
|
AABR07065811.1
|
|
| chr16_-_10941414 | 9.62 |
ENSRNOT00000086627
ENSRNOT00000085414 ENSRNOT00000081631 ENSRNOT00000087521 ENSRNOT00000083623 |
Ldb3
|
LIM domain binding 3 |
| chr13_+_92146586 | 9.55 |
ENSRNOT00000004659
|
Olr1588
|
olfactory receptor 1588 |
| chr4_-_150185044 | 9.28 |
ENSRNOT00000019778
|
Csgalnact2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr15_-_34469350 | 9.15 |
ENSRNOT00000067536
|
Adcy4
|
adenylate cyclase 4 |
| chr14_+_34727623 | 9.04 |
ENSRNOT00000071405
ENSRNOT00000090183 |
Kdr
|
kinase insert domain receptor |
| chr4_+_123118468 | 8.87 |
ENSRNOT00000010895
|
Tmem43
|
transmembrane protein 43 |
| chr2_-_54963448 | 8.77 |
ENSRNOT00000017886
|
Ptger4
|
prostaglandin E receptor 4 |
| chr1_+_81230989 | 8.65 |
ENSRNOT00000077952
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr6_-_51019407 | 8.47 |
ENSRNOT00000011659
|
Gpr22
|
G protein-coupled receptor 22 |
| chr13_-_112099336 | 8.31 |
ENSRNOT00000009158
ENSRNOT00000044161 |
Camk1g
|
calcium/calmodulin-dependent protein kinase IG |
| chr4_+_101909389 | 8.21 |
ENSRNOT00000086458
|
AABR07060963.2
|
|
| chr7_-_71048383 | 8.13 |
ENSRNOT00000005693
|
Gpr182
|
G protein-coupled receptor 182 |
| chr2_+_187977008 | 7.81 |
ENSRNOT00000081531
ENSRNOT00000092951 |
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chrX_+_105573909 | 7.80 |
ENSRNOT00000029664
|
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr6_-_140805551 | 7.58 |
ENSRNOT00000080018
|
AABR07065774.1
|
|
| chr11_-_37993204 | 7.58 |
ENSRNOT00000077050
ENSRNOT00000091614 ENSRNOT00000002683 |
Fam3b
|
family with sequence similarity 3, member B |
| chr7_-_140356209 | 7.51 |
ENSRNOT00000077856
|
Rnd1
|
Rho family GTPase 1 |
| chr14_+_80248140 | 7.41 |
ENSRNOT00000010852
|
Htra3
|
HtrA serine peptidase 3 |
| chr10_-_103919605 | 7.20 |
ENSRNOT00000004718
|
Hid1
|
HID1 domain containing |
| chr6_-_142251976 | 7.20 |
ENSRNOT00000051475
|
AABR07065812.1
|
|
| chr7_+_59200918 | 7.13 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr4_+_7076759 | 7.08 |
ENSRNOT00000066598
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr10_-_57822498 | 6.77 |
ENSRNOT00000037546
|
Nlrp1a
|
NLR family, pyrin domain containing 1A |
| chr1_+_80279706 | 6.74 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr10_-_88754829 | 6.71 |
ENSRNOT00000026354
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr9_-_19749145 | 6.56 |
ENSRNOT00000013956
|
Rcan2
|
regulator of calcineurin 2 |
| chr1_+_266053002 | 6.56 |
ENSRNOT00000026235
|
Nfkb2
|
nuclear factor kappa B subunit 2 |
| chr13_+_83531018 | 6.55 |
ENSRNOT00000004104
|
Gpr161
|
G protein-coupled receptor 161 |
| chr4_-_155421998 | 6.41 |
ENSRNOT00000041552
|
Gdf3
|
growth differentiation factor 3 |
| chr3_-_2411544 | 6.37 |
ENSRNOT00000012406
|
Tor4a
|
torsin family 4, member A |
| chr10_+_13736313 | 6.31 |
ENSRNOT00000079700
|
Abca3
|
ATP binding cassette subfamily A member 3 |
| chr3_+_159995064 | 6.20 |
ENSRNOT00000012606
|
Ttpal
|
alpha tocopherol transfer protein like |
| chr9_+_81655629 | 6.16 |
ENSRNOT00000088679
ENSRNOT00000057472 |
Slc11a1
|
solute carrier family 11 member 1 |
| chr13_+_88265331 | 6.13 |
ENSRNOT00000031190
|
Ccdc190
|
coiled-coil domain containing 190 |
| chr2_+_189423559 | 6.03 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr1_-_72464492 | 6.02 |
ENSRNOT00000068550
|
Nat14
|
N-acetyltransferase 14 |
| chr1_+_213766758 | 5.93 |
ENSRNOT00000005645
|
Ifitm1
|
interferon induced transmembrane protein 1 |
| chr3_+_152857592 | 5.81 |
ENSRNOT00000027445
|
Myl9
|
myosin light chain 9 |
| chr6_-_146195819 | 5.75 |
ENSRNOT00000007625
|
Sp4
|
Sp4 transcription factor |
| chr3_+_168345152 | 5.71 |
ENSRNOT00000017654
|
Dok5
|
docking protein 5 |
| chr13_-_94289333 | 5.65 |
ENSRNOT00000078195
|
Pld5
|
phospholipase D family, member 5 |
| chr15_-_51485692 | 5.61 |
ENSRNOT00000023876
|
Rhobtb2
|
Rho-related BTB domain containing 2 |
| chr7_-_31824064 | 5.61 |
ENSRNOT00000011494
ENSRNOT00000080824 |
Slc25a3
|
solute carrier family 25 member 3 |
| chr1_+_165189985 | 5.51 |
ENSRNOT00000048437
|
Kcne3
|
potassium voltage-gated channel subfamily E regulatory subunit 3 |
| chr10_-_110232843 | 5.50 |
ENSRNOT00000054934
|
Cd7
|
Cd7 molecule |
| chr1_+_215609645 | 5.41 |
ENSRNOT00000076140
ENSRNOT00000027487 |
Tnni2
|
troponin I2, fast skeletal type |
| chr19_-_19376789 | 5.35 |
ENSRNOT00000065103
|
Nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr10_-_56444847 | 5.29 |
ENSRNOT00000056872
ENSRNOT00000092662 |
Nlgn2
|
neuroligin 2 |
| chr1_+_84364368 | 5.25 |
ENSRNOT00000024829
|
RGD1307554
|
similar to CG16812-PA |
| chr10_+_57040267 | 5.19 |
ENSRNOT00000026207
|
Arrb2
|
arrestin, beta 2 |
| chr10_-_82326771 | 5.16 |
ENSRNOT00000004673
|
Acsf2
|
acyl-CoA synthetase family member 2 |
| chr14_-_46153212 | 5.10 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr1_+_257157264 | 5.09 |
ENSRNOT00000067149
ENSRNOT00000079219 |
Plce1
|
phospholipase C, epsilon 1 |
| chr2_-_250981623 | 5.04 |
ENSRNOT00000018435
|
Clca2
|
chloride channel accessory 2 |
| chr1_-_78180216 | 4.89 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr15_-_8989580 | 4.86 |
ENSRNOT00000061402
|
Thrb
|
thyroid hormone receptor beta |
| chr16_-_69965224 | 4.81 |
ENSRNOT00000093000
|
Prrg3
|
proline rich and Gla domain 3 |
| chr5_+_151436464 | 4.80 |
ENSRNOT00000012463
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr14_-_38575785 | 4.74 |
ENSRNOT00000003146
|
Atp10d
|
ATPase phospholipid transporting 10D (putative) |
| chr8_-_71055969 | 4.73 |
ENSRNOT00000075734
|
Ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr5_+_22380334 | 4.71 |
ENSRNOT00000009744
|
Clvs1
|
clavesin 1 |
| chr1_-_221015929 | 4.58 |
ENSRNOT00000028137
|
Sipa1
|
signal-induced proliferation-associated 1 |
| chr3_+_173953869 | 4.56 |
ENSRNOT00000091212
|
Fam217b
|
family with sequence similarity 217, member B |
| chr20_+_46429222 | 4.53 |
ENSRNOT00000076818
|
Foxo3
|
forkhead box O3 |
| chr18_+_30114708 | 4.51 |
ENSRNOT00000027343
|
Pcdha4
|
protocadherin alpha 4 |
| chr18_+_52550739 | 4.48 |
ENSRNOT00000037529
|
Ctxn3
|
cortexin 3 |
| chr9_-_82008620 | 4.46 |
ENSRNOT00000023365
|
Prkag3
|
protein kinase AMP-activated non-catalytic subunit gamma 3 |
| chr4_+_118852062 | 4.37 |
ENSRNOT00000090827
ENSRNOT00000025070 |
Gfpt1
|
glutamine fructose-6-phosphate transaminase 1 |
| chr14_-_78707861 | 4.36 |
ENSRNOT00000091927
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr7_-_141136700 | 4.30 |
ENSRNOT00000091538
|
Bcdin3d
|
BCDIN3 domain containing RNA methyltransferase |
| chr5_+_169338097 | 4.25 |
ENSRNOT00000014035
|
Hes2
|
hes family bHLH transcription factor 2 |
| chr4_+_155653718 | 4.12 |
ENSRNOT00000065419
|
Foxj2
|
forkhead box J2 |
| chr10_+_59539405 | 4.10 |
ENSRNOT00000077107
|
Itgae
|
integrin subunit alpha E |
| chr20_+_4355175 | 4.08 |
ENSRNOT00000000510
|
Gpsm3
|
G-protein signaling modulator 3 |
| chr2_-_187863503 | 4.08 |
ENSRNOT00000093036
ENSRNOT00000026705 ENSRNOT00000082174 |
Lmna
|
lamin A/C |
| chr20_-_3438389 | 4.06 |
ENSRNOT00000001098
ENSRNOT00000090039 |
Flot1
|
flotillin 1 |
| chr14_-_83776863 | 4.02 |
ENSRNOT00000026481
|
Smtn
|
smoothelin |
| chr6_-_141715846 | 4.01 |
ENSRNOT00000044966
|
AABR07065798.1
|
|
| chr3_-_79390956 | 3.94 |
ENSRNOT00000077943
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr4_-_71713063 | 3.91 |
ENSRNOT00000059447
|
Fam131b
|
family with sequence similarity 131, member B |
| chr4_+_153805993 | 3.81 |
ENSRNOT00000056174
|
Usp18
|
ubiquitin specific peptidase 18 |
| chr6_-_141957537 | 3.80 |
ENSRNOT00000090358
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr6_-_132972511 | 3.76 |
ENSRNOT00000082216
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr6_-_61405195 | 3.73 |
ENSRNOT00000008655
|
Lrrn3
|
leucine rich repeat neuronal 3 |
| chr1_+_99505677 | 3.69 |
ENSRNOT00000024645
|
Zfp719
|
zinc finger protein 719 |
| chr1_-_77830399 | 3.61 |
ENSRNOT00000052231
|
LOC100360449
|
ribosomal protein L9-like |
| chr3_-_38090526 | 3.58 |
ENSRNOT00000059430
|
Cacnb4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr1_+_212181374 | 3.56 |
ENSRNOT00000085921
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr6_+_29518416 | 3.53 |
ENSRNOT00000078660
|
AABR07063346.1
|
|
| chr7_+_142905758 | 3.47 |
ENSRNOT00000078663
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chrX_+_39711201 | 3.47 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr10_-_38838272 | 3.44 |
ENSRNOT00000089495
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr12_-_22138382 | 3.40 |
ENSRNOT00000001899
|
Lrch4
|
leucine rich repeats and calponin homology domain containing 4 |
| chr10_-_63491713 | 3.36 |
ENSRNOT00000063941
|
Tusc5
|
tumor suppressor candidate 5 |
| chr20_-_3397039 | 3.24 |
ENSRNOT00000001084
ENSRNOT00000085259 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr13_+_90692666 | 3.23 |
ENSRNOT00000010028
|
Igsf8
|
immunoglobulin superfamily, member 8 |
| chr20_-_11528332 | 3.17 |
ENSRNOT00000038419
|
Tspear
|
thrombospondin-type laminin G domain and EAR repeats |
| chr8_+_102304095 | 3.16 |
ENSRNOT00000011358
|
Slc9a9
|
solute carrier family 9 member A9 |
| chr1_+_192025710 | 3.16 |
ENSRNOT00000077457
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr1_-_197885031 | 3.11 |
ENSRNOT00000071241
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr2_+_78103387 | 3.09 |
ENSRNOT00000063951
|
LOC103689968
|
protein FAM134B |
| chr2_+_210880777 | 3.06 |
ENSRNOT00000026237
|
Gnat2
|
G protein subunit alpha transducin 2 |
| chr1_+_64506735 | 2.92 |
ENSRNOT00000086331
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ebf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.9 | 47.6 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 8.0 | 16.0 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 6.6 | 13.2 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 5.8 | 17.4 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 5.6 | 22.3 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 5.5 | 16.6 | GO:1905204 | response to selenite ion(GO:0072714) negative regulation of connective tissue replacement(GO:1905204) |
| 5.5 | 22.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 5.5 | 16.4 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 5.3 | 16.0 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 4.9 | 14.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 4.7 | 23.4 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 4.5 | 13.5 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 4.5 | 17.9 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 3.9 | 15.7 | GO:0060354 | negative regulation of cell adhesion molecule production(GO:0060354) |
| 3.4 | 10.2 | GO:2001055 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 3.4 | 13.4 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 3.3 | 19.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 3.3 | 9.8 | GO:2000539 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 3.0 | 18.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 2.9 | 14.6 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 2.9 | 8.8 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 2.9 | 43.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 2.8 | 19.6 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 2.7 | 10.8 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 2.6 | 26.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 2.6 | 5.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 2.6 | 10.4 | GO:0030221 | basophil differentiation(GO:0030221) |
| 2.5 | 12.6 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 2.5 | 12.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 2.4 | 14.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 2.4 | 7.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 2.2 | 33.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) auditory receptor cell stereocilium organization(GO:0060088) |
| 2.0 | 17.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 2.0 | 7.8 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 1.9 | 9.3 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 1.8 | 5.5 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 1.8 | 9.0 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 1.8 | 5.4 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) |
| 1.8 | 7.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 1.7 | 18.6 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 1.7 | 6.7 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 1.7 | 45.2 | GO:0014002 | astrocyte development(GO:0014002) |
| 1.6 | 13.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 1.6 | 4.9 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 1.5 | 4.6 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 1.4 | 18.0 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 1.4 | 4.1 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 1.3 | 10.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 1.3 | 8.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 1.2 | 1.2 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 1.1 | 4.4 | GO:1901073 | glucosamine-containing compound biosynthetic process(GO:1901073) |
| 1.1 | 6.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 1.0 | 10.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 1.0 | 4.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 1.0 | 2.9 | GO:0045751 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) regulation of Toll signaling pathway(GO:0008592) negative regulation of Toll signaling pathway(GO:0045751) |
| 1.0 | 45.8 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.9 | 6.4 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.9 | 2.6 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.8 | 12.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.8 | 12.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.8 | 9.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.8 | 4.9 | GO:0007621 | negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.8 | 4.5 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.6 | 38.7 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.6 | 1.9 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.6 | 3.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.6 | 2.5 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.6 | 3.5 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.6 | 6.8 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.5 | 6.6 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.5 | 8.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.5 | 24.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.5 | 19.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.5 | 6.7 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.5 | 2.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.5 | 6.6 | GO:0007614 | short-term memory(GO:0007614) |
| 0.5 | 39.8 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.4 | 3.9 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.4 | 12.5 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.4 | 8.9 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.4 | 16.9 | GO:0048536 | spleen development(GO:0048536) |
| 0.4 | 2.5 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.4 | 2.9 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.4 | 1.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 4.3 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.4 | 2.0 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.4 | 0.8 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.4 | 5.7 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.3 | 2.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 3.2 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 1.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 2.8 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.3 | 0.8 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.3 | 3.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.3 | 2.7 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.3 | 3.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 13.5 | GO:0035904 | aorta development(GO:0035904) |
| 0.3 | 5.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.3 | 4.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 0.7 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.2 | 10.4 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.2 | 3.8 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.2 | 1.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 4.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 9.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 1.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 7.6 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.2 | 5.5 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.2 | 6.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 3.4 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.2 | 1.1 | GO:0021637 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 | 1.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 1.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 2.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 5.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.9 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.1 | 4.5 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.6 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 8.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 5.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.9 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 2.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 4.7 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.6 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.1 | 3.1 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.1 | 2.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 3.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.7 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 2.4 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 7.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.2 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 1.4 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 1.1 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 1.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 1.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.9 | 47.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 10.9 | 43.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 6.2 | 24.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 5.3 | 16.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 5.2 | 20.9 | GO:0045298 | tubulin complex(GO:0045298) |
| 4.4 | 13.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 4.2 | 12.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 4.1 | 12.3 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 2.4 | 7.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 2.1 | 6.3 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 1.7 | 22.6 | GO:0033643 | host cell part(GO:0033643) |
| 1.4 | 6.8 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 1.1 | 23.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 1.1 | 9.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.1 | 16.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.7 | 36.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.7 | 6.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.7 | 10.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.7 | 37.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.7 | 21.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.6 | 9.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.6 | 7.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.5 | 4.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 8.9 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.5 | 23.2 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.4 | 16.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 18.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 9.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 16.7 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 2.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.3 | 4.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 2.6 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 5.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.3 | 5.8 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.3 | 0.9 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.3 | 28.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 7.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 4.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 11.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 18.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 47.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 12.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 11.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 4.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 2.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 2.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 5.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 17.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 58.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.2 | 5.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 5.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 9.5 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 5.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 9.0 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 1.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 6.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 7.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 19.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 7.4 | GO:0005912 | adherens junction(GO:0005912) |
| 0.1 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.7 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 1.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 2.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 9.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 1.6 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 18.4 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 6.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 3.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 13.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.6 | GO:0019012 | virion(GO:0019012) virion part(GO:0044423) |
| 0.0 | 1.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 9.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.4 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.5 | 43.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 11.9 | 47.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 7.8 | 23.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 5.3 | 37.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 5.2 | 20.9 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 4.4 | 13.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 4.2 | 12.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 4.1 | 12.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 3.9 | 19.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 3.5 | 10.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 3.3 | 16.6 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 2.7 | 8.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 2.7 | 10.8 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 2.7 | 18.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 2.6 | 28.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 2.2 | 13.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 2.1 | 12.6 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 1.8 | 16.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 1.7 | 5.2 | GO:0031762 | follicle-stimulating hormone receptor binding(GO:0031762) type 2A serotonin receptor binding(GO:0031826) |
| 1.6 | 6.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.6 | 4.9 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.6 | 11.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.5 | 4.4 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 1.4 | 14.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.3 | 14.8 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 1.3 | 8.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 1.1 | 9.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 1.1 | 15.7 | GO:0015250 | water channel activity(GO:0015250) |
| 1.1 | 13.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.1 | 5.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.1 | 10.8 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 1.0 | 2.9 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.9 | 2.6 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.8 | 2.4 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.8 | 5.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.7 | 2.8 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.7 | 9.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.7 | 17.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.6 | 13.5 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.6 | 12.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.6 | 2.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.5 | 12.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.5 | 3.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.5 | 2.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.5 | 22.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.4 | 4.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.4 | 10.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 2.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.4 | 9.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.4 | 5.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 7.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 10.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 14.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.4 | 7.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 6.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.3 | 3.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 1.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 51.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.3 | 3.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.3 | 4.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 7.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 2.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 5.1 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.3 | 5.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 2.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 5.6 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.2 | 4.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 4.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 5.7 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.2 | 3.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 15.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.2 | 10.7 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.2 | 6.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 1.6 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 16.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 5.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 16.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 1.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 6.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 55.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 2.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 4.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.4 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 2.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.9 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 3.1 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 4.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 6.0 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 26.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 3.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 5.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 12.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.9 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 4.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 3.8 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 16.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 3.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 5.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.1 | GO:0016874 | ligase activity(GO:0016874) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 25.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 1.7 | 23.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.6 | 20.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.3 | 38.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.7 | 13.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.6 | 6.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.6 | 34.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.5 | 10.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.5 | 5.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.5 | 21.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.5 | 17.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.5 | 10.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.5 | 23.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.5 | 9.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 31.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.4 | 24.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 9.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.4 | 20.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.4 | 16.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.4 | 6.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 12.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 16.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 4.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 6.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 5.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 13.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 7.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 2.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 2.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 3.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 6.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 10.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 5.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 6.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 5.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 14.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 3.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 14.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 3.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 5.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 2.1 | 25.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 1.5 | 59.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.3 | 22.9 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 1.3 | 6.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 1.3 | 15.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.2 | 22.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 1.1 | 8.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 1.1 | 18.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.9 | 16.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.8 | 21.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.8 | 23.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.7 | 20.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.7 | 19.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.6 | 8.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.6 | 9.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.6 | 20.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.6 | 14.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.5 | 9.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.5 | 14.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.5 | 4.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.4 | 15.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 5.4 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.4 | 11.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.4 | 9.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.4 | 6.1 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.3 | 7.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 29.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.3 | 21.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 20.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 3.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 2.6 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.2 | 4.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 3.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 39.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 3.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 14.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 5.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 1.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 8.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 5.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 4.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |