Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for E2f6
Z-value: 1.23
Transcription factors associated with E2f6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f6
|
ENSRNOG00000004449 | E2F transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f6 | rn6_v1_chr6_+_42092467_42092467 | -0.29 | 1.5e-07 | Click! |
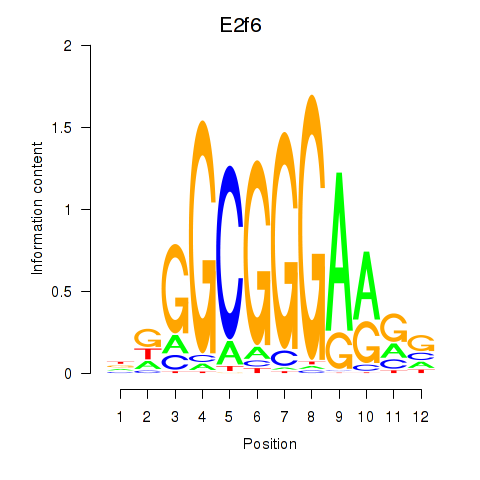
Activity profile of E2f6 motif
Sorted Z-values of E2f6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_71514281 | 36.87 |
ENSRNOT00000022256
|
Ns5atp9
|
NS5A (hepatitis C virus) transactivated protein 9 |
| chr7_-_12741296 | 30.42 |
ENSRNOT00000060648
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr4_+_71740532 | 28.93 |
ENSRNOT00000023537
|
Zyx
|
zyxin |
| chr18_+_51785111 | 28.02 |
ENSRNOT00000019351
|
Lmnb1
|
lamin B1 |
| chr1_+_81230989 | 27.75 |
ENSRNOT00000077952
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr1_+_81230612 | 27.43 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr9_-_10757720 | 27.26 |
ENSRNOT00000083848
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr9_-_10757954 | 26.00 |
ENSRNOT00000075265
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr12_-_21720094 | 24.95 |
ENSRNOT00000032838
|
LOC100910636
|
zinc finger CW-type PWWP domain protein 1-like |
| chr9_-_26932201 | 23.98 |
ENSRNOT00000017081
|
Mcm3
|
minichromosome maintenance complex component 3 |
| chr6_+_43884678 | 23.10 |
ENSRNOT00000091551
|
Rrm2
|
ribonucleotide reductase regulatory subunit M2 |
| chr14_+_108826831 | 21.80 |
ENSRNOT00000083146
ENSRNOT00000009421 |
Bcl11a
|
B-cell CLL/lymphoma 11A |
| chr3_+_59153280 | 21.49 |
ENSRNOT00000002066
|
Cdca7
|
cell division cycle associated 7 |
| chr5_-_24631679 | 21.20 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr20_+_44436403 | 20.62 |
ENSRNOT00000000733
ENSRNOT00000076859 |
Fyn
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr6_+_91463308 | 19.98 |
ENSRNOT00000005595
|
Lrr1
|
leucine rich repeat protein 1 |
| chr2_+_252018597 | 19.75 |
ENSRNOT00000020452
|
Mcoln2
|
mucolipin 2 |
| chr17_+_44520537 | 19.75 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr1_+_144831523 | 19.21 |
ENSRNOT00000039748
|
Mex3b
|
mex-3 RNA binding family member B |
| chr7_-_75421874 | 19.08 |
ENSRNOT00000012775
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr2_-_30780121 | 19.01 |
ENSRNOT00000025129
|
Cenph
|
centromere protein H |
| chr5_+_127404450 | 18.96 |
ENSRNOT00000017575
|
Lrp8
|
LDL receptor related protein 8 |
| chr9_+_81783349 | 18.60 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr1_-_82610350 | 18.29 |
ENSRNOT00000028177
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1 |
| chr17_-_44815995 | 18.22 |
ENSRNOT00000091201
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr7_+_2643288 | 17.59 |
ENSRNOT00000047241
|
Timeless
|
timeless circadian clock |
| chr20_+_3990820 | 17.20 |
ENSRNOT00000000528
|
Psmb8
|
proteasome subunit beta 8 |
| chr9_-_76768770 | 16.87 |
ENSRNOT00000087779
ENSRNOT00000057849 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr16_+_35934970 | 16.85 |
ENSRNOT00000084707
ENSRNOT00000016474 |
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr12_-_46718355 | 16.66 |
ENSRNOT00000030031
|
Rab35
|
RAB35, member RAS oncogene family |
| chr7_+_102586313 | 16.46 |
ENSRNOT00000006188
|
Myc
|
myelocytomatosis oncogene |
| chr17_-_44520240 | 16.02 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr3_-_177201525 | 15.57 |
ENSRNOT00000022451
|
Rgs19
|
regulator of G-protein signaling 19 |
| chr8_-_62424303 | 15.42 |
ENSRNOT00000091223
|
Csk
|
c-src tyrosine kinase |
| chr7_-_63407241 | 15.35 |
ENSRNOT00000024679
|
Tbc1d30
|
TBC1 domain family, member 30 |
| chr12_-_9331195 | 15.35 |
ENSRNOT00000044134
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chr7_+_120125633 | 15.06 |
ENSRNOT00000012480
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr1_+_75356220 | 14.80 |
ENSRNOT00000019799
|
Lig1
|
DNA ligase 1 |
| chr10_-_4910305 | 14.68 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr15_-_24056073 | 14.63 |
ENSRNOT00000015100
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr2_-_207300854 | 14.61 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr1_-_94494980 | 14.56 |
ENSRNOT00000020014
|
Ccne1
|
cyclin E1 |
| chr13_-_110257367 | 14.56 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr16_+_47717951 | 14.35 |
ENSRNOT00000018689
|
Ing2
|
inhibitor of growth family, member 2 |
| chr7_-_31784192 | 14.07 |
ENSRNOT00000010869
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr2_-_45077219 | 14.02 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chr3_+_124068865 | 13.78 |
ENSRNOT00000079264
|
Smox
|
spermine oxidase |
| chr5_+_144581427 | 13.67 |
ENSRNOT00000015227
|
Clspn
|
claspin |
| chr7_+_97984862 | 13.61 |
ENSRNOT00000008508
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr20_-_3419831 | 13.47 |
ENSRNOT00000046798
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr2_-_187820952 | 13.41 |
ENSRNOT00000092762
|
Sema4a
|
semaphorin 4A |
| chr14_-_81725181 | 13.38 |
ENSRNOT00000078252
|
Cfap99
|
cilia and flagella associated protein 99 |
| chr9_+_112293388 | 13.30 |
ENSRNOT00000020767
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr8_+_114867062 | 13.16 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr7_+_144078496 | 13.13 |
ENSRNOT00000055302
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr2_-_206274079 | 13.10 |
ENSRNOT00000056079
|
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr10_-_85049331 | 12.93 |
ENSRNOT00000012538
|
Tbx21
|
T-box 21 |
| chr4_-_81968832 | 12.85 |
ENSRNOT00000016608
|
Skap2
|
src kinase associated phosphoprotein 2 |
| chr7_-_138039984 | 12.76 |
ENSRNOT00000089806
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr16_-_61091169 | 12.65 |
ENSRNOT00000016328
|
Dusp4
|
dual specificity phosphatase 4 |
| chr12_-_38274036 | 12.63 |
ENSRNOT00000063990
|
Kntc1
|
kinetochore associated 1 |
| chr6_-_146195819 | 12.56 |
ENSRNOT00000007625
|
Sp4
|
Sp4 transcription factor |
| chr2_+_113984646 | 12.53 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr7_-_140617721 | 12.51 |
ENSRNOT00000081355
|
Tuba1b
|
tubulin, alpha 1B |
| chr1_+_167538263 | 12.46 |
ENSRNOT00000074058
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr1_-_146029840 | 12.36 |
ENSRNOT00000016455
|
Mesdc1
|
mesoderm development candidate 1 |
| chr19_+_37652969 | 12.32 |
ENSRNOT00000041970
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr7_+_37812831 | 12.27 |
ENSRNOT00000005910
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr2_+_198418691 | 12.25 |
ENSRNOT00000089409
|
Hist1h2bk
|
histone cluster 1, H2bk |
| chr3_-_148722710 | 12.25 |
ENSRNOT00000090919
ENSRNOT00000068592 |
Plagl2
|
PLAG1 like zinc finger 2 |
| chr17_+_6909728 | 12.22 |
ENSRNOT00000061231
|
LOC681410
|
hypothetical protein LOC681410 |
| chr8_-_78655856 | 12.15 |
ENSRNOT00000081185
|
Tcf12
|
transcription factor 12 |
| chr7_-_143863186 | 12.06 |
ENSRNOT00000017096
|
Rarg
|
retinoic acid receptor, gamma |
| chr4_+_70903253 | 12.00 |
ENSRNOT00000019720
|
Ephb6
|
Eph receptor B6 |
| chr8_+_44136496 | 11.95 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr11_-_66695353 | 11.64 |
ENSRNOT00000063995
|
Polq
|
DNA polymerase theta |
| chr9_+_10471742 | 11.51 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chrX_+_113948654 | 11.18 |
ENSRNOT00000068431
|
Tmem164
|
transmembrane protein 164 |
| chrX_+_65226748 | 11.07 |
ENSRNOT00000076181
|
Msn
|
moesin |
| chr10_+_29289203 | 11.07 |
ENSRNOT00000067013
|
Pwwp2a
|
PWWP domain containing 2A |
| chr5_+_150032999 | 11.00 |
ENSRNOT00000013301
|
Srsf4
|
serine and arginine rich splicing factor 4 |
| chr10_+_65606898 | 10.88 |
ENSRNOT00000015091
|
Unc119
|
unc-119 lipid binding chaperone |
| chr8_-_36314811 | 10.87 |
ENSRNOT00000013243
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr1_-_219450451 | 10.67 |
ENSRNOT00000025317
|
Rad9a
|
RAD9 checkpoint clamp component A |
| chr15_+_51756978 | 10.58 |
ENSRNOT00000024067
|
Egr3
|
early growth response 3 |
| chr5_+_128923934 | 10.56 |
ENSRNOT00000064145
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr17_+_72429618 | 10.29 |
ENSRNOT00000026187
|
Gata3
|
GATA binding protein 3 |
| chr10_+_83655460 | 10.20 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr12_+_24978483 | 10.19 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr10_-_40122915 | 10.17 |
ENSRNOT00000063891
|
Cdc42se2
|
CDC42 small effector 2 |
| chr1_+_165382690 | 10.03 |
ENSRNOT00000023802
|
C2cd3
|
C2 calcium-dependent domain containing 3 |
| chr8_+_11888591 | 10.01 |
ENSRNOT00000083967
|
Ccdc82
|
coiled-coil domain containing 82 |
| chr2_+_189430041 | 10.00 |
ENSRNOT00000023567
ENSRNOT00000023605 |
Tpm3
|
tropomyosin 3 |
| chr10_+_11206226 | 9.93 |
ENSRNOT00000006979
|
Tfap4
|
transcription factor AP-4 |
| chr1_+_199412834 | 9.92 |
ENSRNOT00000031891
|
Fus
|
fused in sarcoma RNA binding protein |
| chr1_+_167539036 | 9.90 |
ENSRNOT00000093112
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr2_-_238068694 | 9.86 |
ENSRNOT00000079479
ENSRNOT00000078608 ENSRNOT00000090834 |
Npnt
|
nephronectin |
| chr7_+_123573930 | 9.85 |
ENSRNOT00000066149
|
Fam109b
|
family with sequence similarity 109, member B |
| chr6_-_11274932 | 9.83 |
ENSRNOT00000021538
|
Msh2
|
mutS homolog 2 |
| chr2_-_43999107 | 9.82 |
ENSRNOT00000065473
|
LOC100912399
|
mitogen-activated protein kinase kinase kinase 1-like |
| chr5_-_76756140 | 9.75 |
ENSRNOT00000022107
ENSRNOT00000089251 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr7_+_54247460 | 9.69 |
ENSRNOT00000005361
|
Phlda1
|
pleckstrin homology-like domain, family A, member 1 |
| chr4_-_183426439 | 9.64 |
ENSRNOT00000083310
|
Fam60a
|
family with sequence similarity 60, member A |
| chr3_+_7422820 | 9.57 |
ENSRNOT00000064323
|
Ddx31
|
DEAD-box helicase 31 |
| chr20_+_4392626 | 9.53 |
ENSRNOT00000000495
|
Prrt1
|
proline-rich transmembrane protein 1 |
| chr5_-_152122542 | 9.52 |
ENSRNOT00000068634
ENSRNOT00000077248 |
Rps6ka1
|
ribosomal protein S6 kinase A1 |
| chr12_-_37047628 | 9.52 |
ENSRNOT00000001337
|
Rflna
|
refilin A |
| chr5_-_152198813 | 9.37 |
ENSRNOT00000082953
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr7_-_138039630 | 9.36 |
ENSRNOT00000008138
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr3_+_93920447 | 9.35 |
ENSRNOT00000012625
|
Lmo2
|
LIM domain only 2 |
| chr1_+_198932870 | 9.25 |
ENSRNOT00000055003
|
Fbrs
|
fibrosin |
| chr6_+_99402360 | 9.22 |
ENSRNOT00000078498
|
Zbtb1
|
zinc finger and BTB domain containing 1 |
| chr2_+_212247451 | 9.19 |
ENSRNOT00000027813
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chr3_+_117514444 | 9.19 |
ENSRNOT00000009549
|
Dut
|
deoxyuridine triphosphatase |
| chr7_-_14189688 | 9.08 |
ENSRNOT00000037456
|
Notch3
|
notch 3 |
| chr12_-_24537313 | 8.96 |
ENSRNOT00000001975
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chrX_+_158835811 | 8.93 |
ENSRNOT00000071888
ENSRNOT00000080110 |
Ints6l
|
integrator complex subunit 6 like |
| chr7_+_140315368 | 8.89 |
ENSRNOT00000081206
|
Cacnb3
|
calcium voltage-gated channel auxiliary subunit beta 3 |
| chr7_+_137680530 | 8.85 |
ENSRNOT00000006970
|
Arid2
|
AT-rich interaction domain 2 |
| chr19_+_37576075 | 8.77 |
ENSRNOT00000089022
|
Fam65a
|
family with sequence similarity 65, member A |
| chr19_-_39300893 | 8.71 |
ENSRNOT00000027695
|
Terf2
|
telomeric repeat binding factor 2 |
| chr3_-_146491837 | 8.61 |
ENSRNOT00000061469
|
Vsx1
|
visual system homeobox 1 |
| chr6_-_36819821 | 8.58 |
ENSRNOT00000006499
|
Kcns3
|
potassium voltage-gated channel, modifier subfamily S, member 3 |
| chr8_-_122532070 | 8.52 |
ENSRNOT00000013705
|
Ccr4
|
C-C motif chemokine receptor 4 |
| chr4_+_160020472 | 8.48 |
ENSRNOT00000078802
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr10_-_15228235 | 8.47 |
ENSRNOT00000027121
|
Wdr90
|
WD repeat domain 90 |
| chr9_+_38544398 | 8.46 |
ENSRNOT00000016828
|
Prim2
|
primase (DNA) subunit 2 |
| chr4_-_77347011 | 8.45 |
ENSRNOT00000008149
|
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr1_+_167538744 | 8.34 |
ENSRNOT00000093070
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr4_-_83137527 | 8.31 |
ENSRNOT00000039580
|
Jazf1
|
JAZF zinc finger 1 |
| chr5_+_2632712 | 8.22 |
ENSRNOT00000009431
|
Rpl7
|
ribosomal protein L7 |
| chr6_-_2886465 | 8.13 |
ENSRNOT00000038448
|
Srsf7
|
serine and arginine rich splicing factor 7 |
| chr20_-_46666830 | 8.10 |
ENSRNOT00000000331
|
Cep57l1
|
centrosomal protein 57-like 1 |
| chr8_+_77107536 | 8.08 |
ENSRNOT00000083255
|
Adam10
|
ADAM metallopeptidase domain 10 |
| chr10_-_15098791 | 8.04 |
ENSRNOT00000026139
|
Chtf18
|
chromosome transmission fidelity factor 18 |
| chr3_-_93734282 | 8.00 |
ENSRNOT00000012428
|
Caprin1
|
cell cycle associated protein 1 |
| chr1_-_143751789 | 8.00 |
ENSRNOT00000026754
|
Hdgfrp3
|
hepatoma-derived growth factor, related protein 3 |
| chr13_-_74077783 | 7.98 |
ENSRNOT00000005677
|
Soat1
|
sterol O-acyltransferase 1 |
| chr11_+_82373870 | 7.91 |
ENSRNOT00000002429
|
Tra2b
|
transformer 2 beta homolog (Drosophila) |
| chr10_-_64202380 | 7.85 |
ENSRNOT00000008982
|
Rflnb
|
refilin B |
| chr5_-_169658875 | 7.84 |
ENSRNOT00000015840
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr5_-_144221263 | 7.82 |
ENSRNOT00000013570
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr10_+_82800704 | 7.79 |
ENSRNOT00000089497
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr2_-_104133985 | 7.78 |
ENSRNOT00000088167
ENSRNOT00000067725 |
Pde7a
|
phosphodiesterase 7A |
| chr2_+_188476820 | 7.66 |
ENSRNOT00000027793
|
Clk2
|
CDC-like kinase 2 |
| chr4_+_79557854 | 7.58 |
ENSRNOT00000013145
|
Npy
|
neuropeptide Y |
| chr8_-_115621394 | 7.55 |
ENSRNOT00000018995
|
Rbm15b
|
RNA binding motif protein 15B |
| chr7_-_59514939 | 7.54 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr12_-_25143480 | 7.44 |
ENSRNOT00000001989
|
Rfc2
|
replication factor C subunit 2 |
| chrX_-_31140711 | 7.43 |
ENSRNOT00000082358
|
Fancb
|
Fanconi anemia, complementation group B |
| chr3_-_119619865 | 7.42 |
ENSRNOT00000016172
|
Itpripl1
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 1 |
| chr2_+_226900619 | 7.39 |
ENSRNOT00000019638
|
Pde5a
|
phosphodiesterase 5A |
| chr7_+_54213319 | 7.36 |
ENSRNOT00000005286
|
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr2_-_219458271 | 7.35 |
ENSRNOT00000064735
|
Cdc14a
|
cell division cycle 14A |
| chr4_+_44597123 | 7.33 |
ENSRNOT00000078250
|
Cav1
|
caveolin 1 |
| chr10_+_70689863 | 7.32 |
ENSRNOT00000091122
ENSRNOT00000086987 ENSRNOT00000082030 |
Taf15
|
TATA-box binding protein associated factor 15 |
| chr19_+_25815207 | 7.32 |
ENSRNOT00000003980
|
Lyl1
|
LYL1, basic helix-loop-helix family member |
| chr9_+_82033543 | 7.18 |
ENSRNOT00000023439
|
Wnt6
|
wingless-type MMTV integration site family, member 6 |
| chr10_+_61685241 | 7.17 |
ENSRNOT00000092606
|
Mnt
|
MAX network transcriptional repressor |
| chr19_-_39087880 | 7.17 |
ENSRNOT00000070822
|
Chtf8
|
chromosome transmission fidelity factor 8 |
| chr4_+_79573998 | 7.17 |
ENSRNOT00000074351
|
LOC100912228
|
pro-neuropeptide Y-like |
| chr8_+_78858570 | 7.13 |
ENSRNOT00000089335
|
Zfp280d
|
zinc finger protein 280D |
| chr14_+_83510640 | 7.05 |
ENSRNOT00000089928
ENSRNOT00000082242 ENSRNOT00000025378 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr19_+_39087990 | 7.05 |
ENSRNOT00000027553
|
Utp4
|
UTP4 small subunit processome component |
| chr2_-_88414012 | 6.98 |
ENSRNOT00000014762
|
Lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr7_-_2941122 | 6.95 |
ENSRNOT00000082107
|
Esyt1
|
extended synaptotagmin 1 |
| chr4_-_98593664 | 6.93 |
ENSRNOT00000007927
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr1_-_220644636 | 6.91 |
ENSRNOT00000027632
|
Pacs1
|
phosphofurin acidic cluster sorting protein 1 |
| chr10_+_86819472 | 6.82 |
ENSRNOT00000081424
|
Cdc6
|
cell division cycle 6 |
| chr18_+_36596585 | 6.81 |
ENSRNOT00000036613
|
Rbm27
|
RNA binding motif protein 27 |
| chr10_-_56154548 | 6.80 |
ENSRNOT00000090809
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr5_-_147535240 | 6.80 |
ENSRNOT00000029177
|
Rbbp4
|
RB binding protein 4, chromatin remodeling factor |
| chr1_+_199225100 | 6.80 |
ENSRNOT00000088606
|
Setd1a
|
SET domain containing 1A |
| chr10_+_11786121 | 6.74 |
ENSRNOT00000033919
|
Slx4
|
SLX4 structure-specific endonuclease subunit |
| chr3_-_161498951 | 6.58 |
ENSRNOT00000024214
|
Ncoa5
|
nuclear receptor coactivator 5 |
| chr10_+_108340240 | 6.50 |
ENSRNOT00000077535
|
Ccdc40
|
coiled-coil domain containing 40 |
| chr2_-_38208719 | 6.48 |
ENSRNOT00000019189
|
Kif2a
|
kinesin family member 2A |
| chr6_-_43493816 | 6.45 |
ENSRNOT00000077786
|
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr1_+_154377447 | 6.44 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_+_32058044 | 6.40 |
ENSRNOT00000021898
|
Nkd2
|
naked cuticle homolog 2 |
| chr1_+_266255797 | 6.40 |
ENSRNOT00000027047
|
Trim8
|
tripartite motif-containing 8 |
| chr10_+_67677071 | 6.32 |
ENSRNOT00000007312
|
Rhbdl3
|
rhomboid like 3 |
| chr7_-_81592206 | 6.32 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr1_+_126961389 | 6.30 |
ENSRNOT00000016758
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr6_-_100011226 | 6.27 |
ENSRNOT00000083286
ENSRNOT00000041442 |
Max
|
MYC associated factor X |
| chr9_-_17206994 | 6.27 |
ENSRNOT00000026195
|
Gtpbp2
|
GTP binding protein 2 |
| chr10_+_86819929 | 6.26 |
ENSRNOT00000038228
|
Cdc6
|
cell division cycle 6 |
| chr1_+_225129097 | 6.25 |
ENSRNOT00000026938
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr4_-_115157263 | 6.25 |
ENSRNOT00000015296
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr10_-_13892997 | 6.24 |
ENSRNOT00000004192
|
Traf7
|
TNF receptor associated factor 7 |
| chr4_-_56114254 | 6.22 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr14_+_11198896 | 6.22 |
ENSRNOT00000079767
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr6_-_128149220 | 6.17 |
ENSRNOT00000014204
|
Gsc
|
goosecoid homeobox |
| chr8_-_67869019 | 6.17 |
ENSRNOT00000066009
|
Pias1
|
protein inhibitor of activated STAT, 1 |
| chr4_+_157523110 | 6.14 |
ENSRNOT00000081640
|
Zfp384
|
zinc finger protein 384 |
| chr14_+_19866408 | 6.12 |
ENSRNOT00000060465
|
Adamts3
|
ADAM metallopeptidase with thrombospondin type 1, motif 3 |
| chr3_+_33641616 | 6.01 |
ENSRNOT00000051953
|
Epc2
|
enhancer of polycomb homolog 2 |
| chr1_+_100811755 | 5.97 |
ENSRNOT00000074847
|
Nup62
|
nucleoporin 62 |
| chr20_+_44803885 | 5.92 |
ENSRNOT00000077936
|
Rev3l
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr7_+_70364813 | 5.83 |
ENSRNOT00000084012
ENSRNOT00000031230 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 21.8 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 6.7 | 53.3 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 6.5 | 19.5 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 6.1 | 24.6 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 6.0 | 17.9 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 5.9 | 17.6 | GO:1904976 | cellular response to bleomycin(GO:1904976) |
| 5.8 | 17.4 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 5.7 | 17.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 5.5 | 16.5 | GO:0090094 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) |
| 5.4 | 27.0 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 5.0 | 55.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 4.3 | 4.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 4.0 | 12.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 4.0 | 20.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 3.9 | 63.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 3.7 | 18.6 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 3.7 | 22.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 3.6 | 10.9 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 3.4 | 20.6 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 3.4 | 10.2 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 3.3 | 10.0 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 3.3 | 10.0 | GO:1905247 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 3.3 | 9.9 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 3.2 | 13.0 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 3.2 | 9.7 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 3.2 | 19.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 3.1 | 12.3 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 2.9 | 11.6 | GO:0097681 | double-strand break repair via alternative nonhomologous end joining(GO:0097681) |
| 2.9 | 8.7 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 2.9 | 14.3 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 2.8 | 11.1 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 2.7 | 8.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 2.6 | 18.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 2.6 | 15.4 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 2.6 | 15.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 2.5 | 7.6 | GO:0032902 | nerve growth factor production(GO:0032902) positive regulation of eating behavior(GO:1904000) |
| 2.5 | 22.6 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 2.5 | 14.8 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 2.4 | 14.6 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 2.3 | 13.8 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 2.3 | 9.1 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 2.3 | 18.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 2.1 | 6.4 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 2.1 | 6.4 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 2.1 | 6.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 2.1 | 8.3 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 2.0 | 6.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 2.0 | 6.0 | GO:0046601 | positive regulation of centriole replication(GO:0046601) regulation of mitotic cytokinesis(GO:1902412) |
| 2.0 | 15.9 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 1.9 | 7.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.9 | 13.1 | GO:1904117 | cellular response to vasopressin(GO:1904117) |
| 1.9 | 7.4 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 1.8 | 5.5 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) |
| 1.8 | 5.4 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 1.8 | 7.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 1.8 | 21.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 1.8 | 10.7 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 1.8 | 42.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 1.7 | 24.3 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 1.7 | 6.9 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 1.7 | 20.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 1.7 | 5.1 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 1.7 | 6.7 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) positive regulation of t-circle formation(GO:1904431) |
| 1.7 | 13.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.7 | 5.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 1.7 | 5.0 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 1.6 | 11.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.6 | 4.9 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 1.6 | 9.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 1.6 | 6.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.6 | 6.2 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 1.5 | 10.6 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 1.5 | 4.5 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 1.5 | 8.9 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 1.5 | 7.4 | GO:0060282 | negative regulation of cardiac muscle contraction(GO:0055118) positive regulation of oocyte development(GO:0060282) |
| 1.5 | 5.9 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 1.5 | 13.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 1.4 | 5.8 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 1.4 | 4.3 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 1.4 | 12.9 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 1.4 | 5.7 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 1.4 | 5.7 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 1.4 | 8.5 | GO:0035983 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 1.4 | 2.8 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 1.4 | 5.5 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.4 | 4.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 1.3 | 8.0 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 1.3 | 34.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 1.3 | 5.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 1.3 | 3.8 | GO:2000111 | senescence-associated heterochromatin focus assembly(GO:0035986) positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.2 | 9.9 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 1.2 | 13.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 1.2 | 3.7 | GO:1903588 | blood vessel lumenization(GO:0072554) negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.2 | 13.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 1.2 | 15.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 1.2 | 3.5 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 1.1 | 9.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 1.1 | 12.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.1 | 12.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 1.1 | 3.3 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 1.1 | 8.8 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 1.1 | 17.5 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 1.1 | 18.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 1.0 | 4.0 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 1.0 | 4.0 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 1.0 | 2.9 | GO:1904954 | regulation of collateral sprouting in absence of injury(GO:0048696) Spemann organizer formation(GO:0060061) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 1.0 | 4.8 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.9 | 12.3 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.9 | 4.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.9 | 2.8 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.9 | 3.7 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.9 | 6.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.9 | 7.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.9 | 8.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.9 | 4.5 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.9 | 13.6 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.9 | 2.7 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 0.9 | 23.0 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.8 | 6.8 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.8 | 8.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.8 | 4.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.8 | 2.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.8 | 7.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.8 | 9.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.8 | 3.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.8 | 13.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.8 | 3.0 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.7 | 12.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.7 | 7.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.7 | 10.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.7 | 3.4 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.7 | 12.2 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.7 | 18.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.7 | 2.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.7 | 2.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.7 | 2.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.7 | 3.9 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.6 | 7.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.6 | 1.9 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.6 | 5.6 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.6 | 4.8 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.6 | 3.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.6 | 2.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.6 | 8.3 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.5 | 2.2 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.5 | 3.8 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.5 | 2.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.5 | 2.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.5 | 1.5 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.5 | 3.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.5 | 16.3 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.5 | 10.9 | GO:0097503 | sialylation(GO:0097503) |
| 0.5 | 7.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.5 | 6.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.5 | 4.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.5 | 3.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.5 | 4.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.5 | 1.4 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.5 | 5.9 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.4 | 6.7 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.4 | 4.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.4 | 11.5 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.4 | 3.0 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.4 | 7.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.4 | 6.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.4 | 2.9 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.4 | 4.8 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.4 | 3.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 5.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.4 | 5.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.4 | 3.4 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.4 | 11.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.4 | 10.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.4 | 2.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 2.2 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.4 | 1.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.4 | 11.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.4 | 5.0 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 0.4 | 6.4 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.4 | 4.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.3 | 1.7 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.3 | 3.7 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 | 1.7 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 2.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.3 | 8.5 | GO:0002507 | tolerance induction(GO:0002507) regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) |
| 0.3 | 8.0 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.3 | 18.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.3 | 18.0 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.3 | 1.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 | 5.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.3 | 1.2 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.3 | 4.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.3 | 1.4 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.3 | 2.3 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.3 | 2.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.3 | 1.7 | GO:0006863 | purine nucleobase transport(GO:0006863) nucleobase transport(GO:0015851) |
| 0.3 | 15.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.3 | 3.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 4.7 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.3 | 3.1 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.3 | 2.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.3 | 3.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 4.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 1.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.2 | 7.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.2 | 4.8 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.2 | 2.0 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.2 | 0.9 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 3.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 8.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 4.5 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.2 | 8.5 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.2 | 18.0 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.2 | 1.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 1.6 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 1.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 3.2 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.2 | 1.6 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.2 | 6.3 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 7.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 1.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.6 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.2 | 1.6 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 1.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.2 | 4.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 2.7 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.2 | 1.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 12.3 | GO:0007569 | cell aging(GO:0007569) |
| 0.2 | 3.4 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.1 | 6.5 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.1 | 6.9 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 2.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 6.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 2.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 2.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 2.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.1 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 5.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 1.0 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 3.3 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 3.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 2.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 7.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 1.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 2.6 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 3.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 7.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 7.1 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.1 | 0.5 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.3 | GO:0060743 | prostate gland stromal morphogenesis(GO:0060741) epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 2.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 11.3 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 3.6 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 4.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 6.2 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 3.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.2 | GO:0014826 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.1 | 1.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 18.1 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.1 | 1.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 1.1 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.1 | 1.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 8.2 | GO:0042113 | B cell activation(GO:0042113) |
| 0.1 | 4.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.7 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 1.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.3 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.2 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 6.9 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 4.0 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 4.1 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 0.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.0 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.5 | 53.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 4.3 | 13.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 4.2 | 12.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 3.5 | 17.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 3.5 | 28.0 | GO:0005638 | lamin filament(GO:0005638) |
| 2.9 | 17.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 2.8 | 14.1 | GO:0043293 | apoptosome(GO:0043293) |
| 2.7 | 13.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 2.5 | 22.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 2.4 | 9.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 2.3 | 6.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 2.2 | 6.7 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 2.2 | 23.9 | GO:0042555 | MCM complex(GO:0042555) |
| 2.1 | 14.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 2.0 | 14.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 2.0 | 10.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 2.0 | 2.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 2.0 | 92.4 | GO:0000791 | euchromatin(GO:0000791) |
| 2.0 | 9.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.8 | 10.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 1.7 | 13.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.6 | 20.9 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 1.5 | 4.5 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 1.4 | 10.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.4 | 7.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 1.4 | 11.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.4 | 20.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 1.3 | 5.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 1.3 | 15.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.3 | 16.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.2 | 12.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 1.2 | 10.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.2 | 5.9 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 1.1 | 3.3 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 1.1 | 4.3 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 1.0 | 18.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 1.0 | 5.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 1.0 | 8.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.0 | 6.0 | GO:0005642 | annulate lamellae(GO:0005642) Flemming body(GO:0090543) |
| 1.0 | 4.8 | GO:0016342 | catenin complex(GO:0016342) |
| 1.0 | 3.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.9 | 4.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.9 | 2.8 | GO:0000811 | GINS complex(GO:0000811) |
| 0.9 | 3.7 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.9 | 16.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.9 | 2.8 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.9 | 4.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.8 | 22.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.8 | 17.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.8 | 6.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.8 | 5.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.8 | 2.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.7 | 9.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.7 | 8.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.7 | 4.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.7 | 24.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.7 | 64.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.7 | 6.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.6 | 5.7 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.6 | 11.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.6 | 0.6 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.6 | 2.9 | GO:0097361 | CIA complex(GO:0097361) |
| 0.6 | 7.0 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.6 | 11.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.5 | 2.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 4.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.5 | 4.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.5 | 7.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 43.9 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.5 | 45.7 | GO:0016605 | PML body(GO:0016605) |
| 0.5 | 7.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.5 | 10.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.5 | 8.7 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.5 | 4.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.4 | 14.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.4 | 2.6 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.4 | 1.3 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.4 | 4.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.4 | 4.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.4 | 38.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.4 | 18.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.4 | 9.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.4 | 9.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 2.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.4 | 2.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 3.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 2.1 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.3 | 22.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 8.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.3 | 3.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.3 | 5.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.3 | 12.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.3 | 13.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 12.2 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 5.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.2 | 19.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.2 | 1.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 4.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.2 | 1.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 9.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 18.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 6.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 23.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.2 | 2.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 3.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 6.8 | GO:0030286 | dynein complex(GO:0030286) |
| 0.2 | 18.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.2 | 4.1 | GO:0044447 | axoneme part(GO:0044447) |
| 0.2 | 36.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.2 | 1.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 3.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 0.5 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 10.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 4.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 6.5 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 32.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 8.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 5.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 6.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 48.0 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 2.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 3.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 3.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 4.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 1.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.1 | GO:0035770 | ribonucleoprotein granule(GO:0035770) |
| 0.0 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 4.3 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.9 | GO:0005819 | spindle(GO:0005819) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.7 | 58.7 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 9.0 | 53.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 7.9 | 55.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 5.5 | 22.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 5.2 | 20.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 4.6 | 13.8 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 4.3 | 13.0 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 4.2 | 12.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 3.4 | 10.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 2.8 | 14.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 2.7 | 13.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 2.7 | 10.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 2.4 | 9.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 2.3 | 34.3 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 2.1 | 19.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 2.1 | 10.6 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 2.1 | 2.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 1.9 | 13.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 1.7 | 5.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 1.6 | 14.8 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.6 | 4.8 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.6 | 11.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 1.6 | 25.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 1.6 | 6.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 1.5 | 6.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 1.5 | 20.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 1.5 | 6.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.5 | 4.5 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 1.5 | 11.9 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.5 | 6.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 1.5 | 17.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 1.4 | 17.8 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 1.3 | 6.7 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 1.3 | 14.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 1.3 | 7.8 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 1.2 | 5.0 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 1.2 | 5.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.2 | 15.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 1.1 | 10.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 1.1 | 4.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 1.0 | 6.2 | GO:0070728 | leucine binding(GO:0070728) |
| 1.0 | 7.9 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 1.0 | 4.8 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 1.0 | 3.8 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.9 | 3.8 | GO:0001156 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.9 | 5.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.9 | 4.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.9 | 4.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.9 | 15.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.9 | 3.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.9 | 19.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.8 | 8.5 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.8 | 5.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.8 | 3.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.8 | 8.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.8 | 15.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.7 | 5.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.7 | 2.9 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.7 | 18.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.7 | 13.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.7 | 23.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.7 | 6.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.7 | 3.5 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.7 | 4.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.7 | 26.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.7 | 15.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.6 | 16.6 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.6 | 17.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.6 | 5.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.6 | 8.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.6 | 18.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.6 | 3.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.6 | 4.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.6 | 12.0 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.6 | 2.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.6 | 3.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.6 | 10.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.6 | 28.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.6 | 11.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.6 | 6.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.6 | 8.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.6 | 3.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.6 | 2.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.6 | 7.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.6 | 17.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.6 | 12.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.6 | 5.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.6 | 4.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.6 | 3.3 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.5 | 5.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.5 | 2.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.5 | 13.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.5 | 13.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.5 | 3.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.5 | 1.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.5 | 4.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.5 | 2.6 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.5 | 12.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.5 | 19.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.5 | 4.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.4 | 2.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.4 | 1.7 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.4 | 8.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.4 | 4.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 2.8 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.4 | 2.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.4 | 3.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.4 | 4.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.4 | 14.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 1.8 | GO:0005111 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.4 | 11.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 7.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.3 | 25.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.3 | 2.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.3 | 3.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.3 | 3.7 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 16.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.3 | 53.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.3 | 3.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.3 | 4.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.3 | 7.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.3 | 1.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 8.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.3 | 6.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 9.9 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.3 | 5.1 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.3 | 11.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.3 | 4.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.3 | 2.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 2.9 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.2 | 1.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 16.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 6.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 1.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 6.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 2.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 1.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.2 | 2.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 2.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 2.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 3.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 1.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 5.8 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 7.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 18.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 2.9 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 3.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 3.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 5.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.9 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 5.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 2.9 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 5.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 10.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 10.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 3.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 3.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 3.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 2.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 6.9 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 6.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 6.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 10.0 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 2.9 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 43.6 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 1.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 49.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 1.5 | 134.3 | PID E2F PATHWAY | E2F transcription factor network |
| 1.4 | 15.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 1.2 | 49.4 | PID ATR PATHWAY | ATR signaling pathway |
| 1.0 | 3.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.9 | 14.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.9 | 27.9 | PID ATM PATHWAY | ATM pathway |
| 0.9 | 23.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.8 | 14.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.7 | 26.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.7 | 20.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.7 | 11.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.6 | 14.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.6 | 16.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.6 | 33.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.6 | 15.4 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.5 | 12.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.5 | 2.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 5.6 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.5 | 12.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.4 | 9.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.4 | 21.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.4 | 10.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 2.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 7.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 5.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 9.8 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 10.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 9.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 9.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 4.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 11.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 19.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 4.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.2 | 2.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 4.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 3.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 4.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 5.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 3.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 4.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 3.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | PID ENDOTHELIN PATHWAY | Endothelins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 26.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 2.3 | 50.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 2.0 | 23.5 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 1.9 | 28.1 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 1.6 | 12.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 1.5 | 15.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 1.5 | 7.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 1.5 | 20.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 1.5 | 37.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 1.4 | 5.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.2 | 22.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 1.2 | 10.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 1.1 | 19.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 1.1 | 8.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 1.1 | 25.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 1.1 | 12.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 1.1 | 19.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.9 | 16.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.9 | 4.5 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.9 | 6.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.8 | 20.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.8 | 13.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.8 | 18.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.8 | 22.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.7 | 4.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.7 | 30.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.7 | 12.0 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.7 | 12.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.7 | 10.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.6 | 16.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.6 | 49.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.6 | 13.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.6 | 11.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.6 | 18.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.6 | 22.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.6 | 7.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.6 | 8.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.5 | 2.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 19.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.5 | 52.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.5 | 7.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.5 | 7.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.5 | 11.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.5 | 7.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.5 | 7.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.5 | 6.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.5 | 4.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 5.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 7.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.4 | 12.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 17.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.4 | 7.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.4 | 9.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.4 | 5.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 9.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.3 | 8.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 3.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 3.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 3.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.3 | 3.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 16.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 4.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 4.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 1.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 6.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.2 | 11.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 3.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 5.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 8.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 8.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 19.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.2 | 4.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 5.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.7 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.1 | 4.8 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 3.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 16.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.4 | REACTOME MITOTIC G1 G1 S PHASES | Genes involved in Mitotic G1-G1/S phases |
| 0.1 | 1.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 4.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 6.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 3.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 3.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |