Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for E2f3
Z-value: 0.82
Transcription factors associated with E2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f3
|
ENSRNOG00000029273 | E2F transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f3 | rn6_v1_chr17_+_36334147_36334147 | 0.52 | 3.0e-23 | Click! |
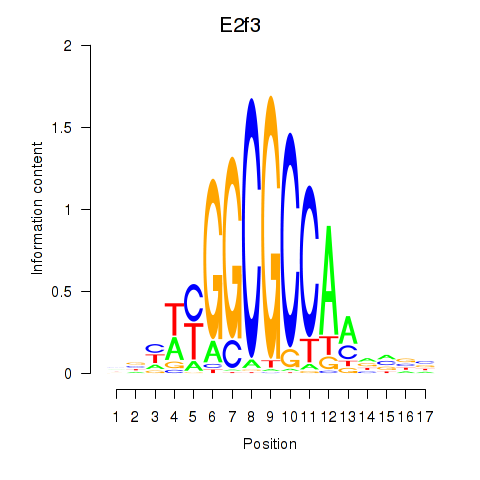
Activity profile of E2f3 motif
Sorted Z-values of E2f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_121302740 | 18.72 |
ENSRNOT00000067032
|
Rpl3
|
ribosomal protein L3 |
| chr13_-_45068077 | 18.56 |
ENSRNOT00000004969
|
Mcm6
|
minichromosome maintenance complex component 6 |
| chr3_+_100366168 | 14.44 |
ENSRNOT00000006689
|
Kif18a
|
kinesin family member 18A |
| chr12_-_51877624 | 14.28 |
ENSRNOT00000056800
|
Chek2
|
checkpoint kinase 2 |
| chr8_-_13262729 | 14.01 |
ENSRNOT00000012176
|
Fut4
|
fucosyltransferase 4 |
| chr6_-_92760018 | 13.68 |
ENSRNOT00000009560
|
Trim9
|
tripartite motif-containing 9 |
| chr14_+_91783514 | 13.59 |
ENSRNOT00000080753
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr10_-_108196217 | 13.07 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr10_-_89454681 | 13.03 |
ENSRNOT00000028109
|
Brca1
|
BRCA1, DNA repair associated |
| chr3_+_146695344 | 13.00 |
ENSRNOT00000010955
|
Gins1
|
GINS complex subunit 1 |
| chr2_+_49682754 | 12.94 |
ENSRNOT00000079907
ENSRNOT00000085576 |
Emb
|
embigin |
| chr19_+_55381565 | 12.85 |
ENSRNOT00000018923
|
Cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr6_-_91518996 | 12.84 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr2_-_198412350 | 12.19 |
ENSRNOT00000040210
|
LOC100912489
|
histone H4-like |
| chr2_+_149214265 | 11.90 |
ENSRNOT00000084020
|
Med12l
|
mediator complex subunit 12-like |
| chr4_+_56981283 | 11.87 |
ENSRNOT00000010989
|
Tspan33
|
tetraspanin 33 |
| chr2_-_157759819 | 11.52 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr10_+_59765258 | 11.17 |
ENSRNOT00000026351
|
Shpk
|
sedoheptulokinase |
| chr11_+_61321459 | 10.95 |
ENSRNOT00000002759
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr18_-_11858744 | 10.68 |
ENSRNOT00000061417
ENSRNOT00000082891 |
Dsc2
|
desmocollin 2 |
| chr13_-_110257367 | 10.06 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chrX_+_156963870 | 9.94 |
ENSRNOT00000077109
|
Pdzd4
|
PDZ domain containing 4 |
| chr3_-_170040953 | 9.83 |
ENSRNOT00000005866
|
Cbln4
|
cerebellin 4 precursor |
| chr3_+_138715570 | 9.49 |
ENSRNOT00000064723
|
Sec23b
|
Sec23 homolog B, coat complex II component |
| chr8_-_21968415 | 9.25 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr1_-_42467586 | 8.77 |
ENSRNOT00000029416
|
Fbxo5
|
F-box protein 5 |
| chr9_+_10471742 | 8.74 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chrX_-_62903530 | 8.56 |
ENSRNOT00000076652
ENSRNOT00000017370 |
Pdk3
|
pyruvate dehydrogenase kinase 3 |
| chr19_+_52521809 | 8.40 |
ENSRNOT00000081019
|
Klhl36
|
kelch-like family member 36 |
| chr9_+_112293388 | 8.25 |
ENSRNOT00000020767
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr7_-_124491004 | 8.03 |
ENSRNOT00000037710
|
Ttll12
|
tubulin tyrosine ligase like 12 |
| chr5_+_79179417 | 7.78 |
ENSRNOT00000010454
|
Orm1
|
orosomucoid 1 |
| chr1_-_261051498 | 7.67 |
ENSRNOT00000071417
|
Arhgap19
|
Rho GTPase activating protein 19 |
| chr3_-_23474170 | 7.64 |
ENSRNOT00000039410
|
Scai
|
suppressor of cancer cell invasion |
| chr8_+_118378059 | 7.59 |
ENSRNOT00000043247
|
AABR07071482.1
|
|
| chr8_-_118378460 | 7.23 |
ENSRNOT00000047247
|
RGD1563705
|
similar to ribosomal protein S23 |
| chr2_-_23256158 | 7.15 |
ENSRNOT00000015336
|
Bhmt
|
betaine-homocysteine S-methyltransferase |
| chr4_-_71227872 | 7.12 |
ENSRNOT00000050392
|
Tcaf2
|
TRPM8 channel-associated factor 2 |
| chr4_-_62438958 | 6.86 |
ENSRNOT00000014010
|
Wdr91
|
WD repeat domain 91 |
| chr1_-_141188031 | 6.85 |
ENSRNOT00000044567
|
Polg
|
DNA polymerase gamma, catalytic subunit |
| chr10_+_4578469 | 6.72 |
ENSRNOT00000003332
|
Txndc11
|
thioredoxin domain containing 11 |
| chr7_+_80351774 | 6.70 |
ENSRNOT00000081948
|
Oxr1
|
oxidation resistance 1 |
| chrX_+_71324365 | 6.61 |
ENSRNOT00000004911
|
Nono
|
non-POU domain containing, octamer-binding |
| chr10_-_53037816 | 6.61 |
ENSRNOT00000057509
|
Shisa6
|
shisa family member 6 |
| chr17_-_9837293 | 6.46 |
ENSRNOT00000091320
ENSRNOT00000022134 |
Prelid1
|
PRELI domain containing 1 |
| chr1_+_31835000 | 6.39 |
ENSRNOT00000020780
|
Cep72
|
centrosomal protein 72 |
| chr9_-_103207190 | 6.26 |
ENSRNOT00000026010
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr2_+_182006242 | 6.19 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr11_-_51202703 | 6.10 |
ENSRNOT00000002719
ENSRNOT00000077220 |
Cblb
|
Cbl proto-oncogene B |
| chr17_-_20364714 | 5.99 |
ENSRNOT00000070962
|
Jarid2
|
jumonji and AT-rich interaction domain containing 2 |
| chr3_-_150603082 | 5.97 |
ENSRNOT00000024310
|
Ahcy
|
adenosylhomocysteinase |
| chr19_+_24329544 | 5.94 |
ENSRNOT00000080934
|
Tbc1d9
|
TBC1 domain family member 9 |
| chr18_+_3163214 | 5.86 |
ENSRNOT00000017291
|
Rbbp8
|
RB binding protein 8, endonuclease |
| chr3_-_10161989 | 5.59 |
ENSRNOT00000012312
|
Exosc2
|
exosome component 2 |
| chr7_+_143754892 | 5.59 |
ENSRNOT00000085896
|
Soat2
|
sterol O-acyltransferase 2 |
| chr20_-_6864571 | 5.54 |
ENSRNOT00000093253
|
Ppil1
|
peptidylprolyl isomerase like 1 |
| chr12_+_43940929 | 5.47 |
ENSRNOT00000001486
|
Rnft2
|
ring finger protein, transmembrane 2 |
| chr8_+_122197027 | 5.46 |
ENSRNOT00000013050
|
Ubp1
|
upstream binding protein 1 (LBP-1a) |
| chr8_-_115621394 | 5.40 |
ENSRNOT00000018995
|
Rbm15b
|
RNA binding motif protein 15B |
| chr2_+_4942775 | 5.35 |
ENSRNOT00000093548
ENSRNOT00000093741 |
Fam172a
|
family with sequence similarity 172, member A |
| chr3_-_94657377 | 5.34 |
ENSRNOT00000077484
|
Tcp11l1
|
t-complex 11 like 1 |
| chr12_+_22434814 | 5.31 |
ENSRNOT00000076829
|
Slc12a9
|
solute carrier family 12, member 9 |
| chr9_-_1984654 | 5.19 |
ENSRNOT00000018146
|
Plcl2
|
phospholipase C-like 2 |
| chr15_+_45422010 | 5.12 |
ENSRNOT00000012231
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr2_+_95008311 | 5.12 |
ENSRNOT00000077270
|
Tpd52
|
tumor protein D52 |
| chr2_+_95008477 | 5.09 |
ENSRNOT00000015327
|
Tpd52
|
tumor protein D52 |
| chr12_-_47793534 | 5.05 |
ENSRNOT00000001588
|
Fam222a
|
family with sequence similarity 222, member A |
| chr7_-_34485034 | 5.01 |
ENSRNOT00000007351
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr8_+_7128656 | 5.00 |
ENSRNOT00000038313
|
Pgr
|
progesterone receptor |
| chr7_-_111722964 | 5.00 |
ENSRNOT00000041802
|
Rps19l1
|
ribosomal protein S19-like 1 |
| chr18_+_3162543 | 4.97 |
ENSRNOT00000078615
|
Rbbp8
|
RB binding protein 8, endonuclease |
| chr5_-_159602251 | 4.96 |
ENSRNOT00000011394
|
Necap2
|
NECAP endocytosis associated 2 |
| chr3_+_22640545 | 4.81 |
ENSRNOT00000064507
ENSRNOT00000014452 |
Lhx2
|
LIM homeobox 2 |
| chr1_-_112947399 | 4.79 |
ENSRNOT00000093306
ENSRNOT00000093259 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr13_+_109713489 | 4.72 |
ENSRNOT00000004962
|
Batf3
|
basic leucine zipper ATF-like transcription factor 3 |
| chr6_-_135049728 | 4.69 |
ENSRNOT00000009556
|
Hsp90aa1
|
heat shock protein 90 alpha family class A member 1 |
| chrX_-_131343853 | 4.69 |
ENSRNOT00000038653
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr20_-_6864387 | 4.65 |
ENSRNOT00000068527
|
Ppil1
|
peptidylprolyl isomerase like 1 |
| chr20_+_2501252 | 4.62 |
ENSRNOT00000079307
ENSRNOT00000084559 |
Trim39
|
tripartite motif-containing 39 |
| chr9_+_111327402 | 4.60 |
ENSRNOT00000045749
|
RGD1562136
|
similar to D1Ertd622e protein |
| chr14_+_58877806 | 4.53 |
ENSRNOT00000051559
|
RGD1562755
|
similar to 60S ribosomal protein L23a |
| chr1_-_112947162 | 4.53 |
ENSRNOT00000014573
ENSRNOT00000083894 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr15_+_27739251 | 4.48 |
ENSRNOT00000011840
|
Parp2
|
poly (ADP-ribose) polymerase 2 |
| chr9_-_46309451 | 4.46 |
ENSRNOT00000018684
|
Rnf149
|
ring finger protein 149 |
| chr20_+_48504264 | 4.46 |
ENSRNOT00000087740
|
Cdc40
|
cell division cycle 40 |
| chr7_-_55604403 | 4.45 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr4_-_177331874 | 4.44 |
ENSRNOT00000065387
ENSRNOT00000091099 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr2_+_128675814 | 4.44 |
ENSRNOT00000058366
|
RGD1359508
|
similar to protein C33A12.3 |
| chr13_+_74154835 | 4.44 |
ENSRNOT00000059524
|
Abl2
|
ABL proto-oncogene 2, non-receptor tyrosine kinase |
| chr10_+_57057608 | 4.27 |
ENSRNOT00000085810
ENSRNOT00000026335 |
Med11
|
mediator complex subunit 11 |
| chr1_-_128341240 | 4.16 |
ENSRNOT00000070864
ENSRNOT00000072915 |
Mef2a
|
myocyte enhancer factor 2a |
| chr3_-_124884570 | 4.16 |
ENSRNOT00000028887
|
Pcna
|
proliferating cell nuclear antigen |
| chr1_-_101095594 | 4.15 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr12_-_14175945 | 4.12 |
ENSRNOT00000001469
|
Ap5z1
|
adaptor-related protein complex 5, zeta 1 subunit |
| chr10_-_72142533 | 4.11 |
ENSRNOT00000030885
|
Mrm1
|
mitochondrial rRNA methyltransferase 1 |
| chr18_+_35489274 | 4.07 |
ENSRNOT00000073952
ENSRNOT00000076060 |
Dcp2
|
decapping mRNA 2 |
| chr10_-_15577977 | 4.03 |
ENSRNOT00000052292
|
Hba-a3
|
hemoglobin alpha, adult chain 3 |
| chr3_-_146690375 | 4.03 |
ENSRNOT00000010641
|
Abhd12
|
abhydrolase domain containing 12 |
| chr13_-_44345735 | 3.99 |
ENSRNOT00000005006
|
Tmem163
|
transmembrane protein 163 |
| chr12_+_24651314 | 3.98 |
ENSRNOT00000077016
ENSRNOT00000071569 |
Vps37d
|
VPS37D, ESCRT-I subunit |
| chr20_-_34929965 | 3.96 |
ENSRNOT00000004499
|
Mcm9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chrX_+_71540895 | 3.90 |
ENSRNOT00000004692
ENSRNOT00000082967 |
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr1_-_141111400 | 3.88 |
ENSRNOT00000022962
|
Rlbp1
|
retinaldehyde binding protein 1 |
| chr5_-_63192025 | 3.86 |
ENSRNOT00000008913
|
Alg2
|
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chrX_+_74200972 | 3.85 |
ENSRNOT00000076956
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr1_-_219863926 | 3.85 |
ENSRNOT00000026454
ENSRNOT00000066455 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr7_-_120780641 | 3.82 |
ENSRNOT00000076164
|
Ddx17
|
DEAD-box helicase 17 |
| chr10_-_67285617 | 3.81 |
ENSRNOT00000019044
|
Utp6
|
UTP6 small subunit processome component |
| chr1_-_84008293 | 3.81 |
ENSRNOT00000002053
|
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chrX_-_62698830 | 3.78 |
ENSRNOT00000076359
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr19_-_59384297 | 3.77 |
ENSRNOT00000077516
|
Tarbp1
|
TAR RNA binding protein 1 |
| chr8_-_48619592 | 3.74 |
ENSRNOT00000012534
|
Abcg4
|
ATP binding cassette subfamily G member 4 |
| chrX_+_74205842 | 3.73 |
ENSRNOT00000077003
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr1_+_165382690 | 3.71 |
ENSRNOT00000023802
|
C2cd3
|
C2 calcium-dependent domain containing 3 |
| chr4_-_180505916 | 3.69 |
ENSRNOT00000086465
|
AABR07062512.1
|
|
| chr9_-_98536581 | 3.69 |
ENSRNOT00000027295
|
Ilkap
|
integrin-linked kinase-associated serine/threonine phosphatase |
| chr6_-_15191660 | 3.65 |
ENSRNOT00000092654
|
Nrxn1
|
neurexin 1 |
| chr18_-_38088457 | 3.57 |
ENSRNOT00000077814
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr13_-_73819896 | 3.56 |
ENSRNOT00000036392
|
Fam163a
|
family with sequence similarity 163, member A |
| chr7_+_73588163 | 3.49 |
ENSRNOT00000015707
|
Kcns2
|
potassium voltage-gated channel, modifier subfamily S, member 2 |
| chr14_+_75852060 | 3.41 |
ENSRNOT00000075975
|
Hs3st1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr10_-_2037891 | 3.38 |
ENSRNOT00000004563
|
Ercc4
|
ERCC excision repair 4, endonuclease catalytic subunit |
| chr14_-_44375804 | 3.36 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr20_+_29831314 | 3.34 |
ENSRNOT00000000696
|
Psap
|
prosaposin |
| chr11_+_61748883 | 3.32 |
ENSRNOT00000093552
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr10_-_13542077 | 3.29 |
ENSRNOT00000008736
|
Atp6v0c
|
ATPase H+ transporting V0 subunit C |
| chr15_-_40545824 | 3.27 |
ENSRNOT00000017204
|
Nup58
|
nucleoporin 58 |
| chrX_-_1704033 | 3.24 |
ENSRNOT00000051956
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr5_+_172648950 | 3.23 |
ENSRNOT00000055361
|
Faap20
|
Fanconi anemia core complex associated protein 20 |
| chr15_-_24374753 | 3.22 |
ENSRNOT00000016274
|
Atg14
|
autophagy related 14 |
| chr20_-_10407554 | 3.19 |
ENSRNOT00000074081
|
U2af1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr10_+_975697 | 3.09 |
ENSRNOT00000089404
|
AC117889.1
|
|
| chr1_-_72329856 | 3.08 |
ENSRNOT00000021391
|
U2af2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr15_-_34338956 | 3.07 |
ENSRNOT00000026914
|
Mdp1
|
magnesium-dependent phosphatase 1 |
| chr8_+_118525682 | 2.99 |
ENSRNOT00000028288
|
Elp6
|
elongator acetyltransferase complex subunit 6 |
| chr19_-_56731372 | 2.98 |
ENSRNOT00000024182
|
Nup133
|
nucleoporin 133 |
| chr2_-_30664163 | 2.96 |
ENSRNOT00000024801
|
Rad17
|
RAD17 checkpoint clamp loader component |
| chrX_+_157150655 | 2.90 |
ENSRNOT00000090795
|
Pnck
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr7_-_11406771 | 2.90 |
ENSRNOT00000047450
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr10_+_40553180 | 2.88 |
ENSRNOT00000087763
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr18_+_24584900 | 2.85 |
ENSRNOT00000017075
|
Wdr33
|
WD repeat domain 33 |
| chr15_-_30147793 | 2.84 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr1_+_220746387 | 2.82 |
ENSRNOT00000027753
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr4_+_157438605 | 2.80 |
ENSRNOT00000079988
|
AC115420.4
|
|
| chr17_-_17947777 | 2.73 |
ENSRNOT00000036876
|
Rnf144b
|
ring finger protein 144B |
| chr10_-_71441389 | 2.73 |
ENSRNOT00000003699
|
Tada2a
|
transcriptional adaptor 2A |
| chr5_+_47853818 | 2.61 |
ENSRNOT00000009228
ENSRNOT00000079656 |
Casp8ap2
|
caspase 8 associated protein 2 |
| chr10_+_14122878 | 2.57 |
ENSRNOT00000052008
|
Hs3st6
|
heparan sulfate-glucosamine 3-sulfotransferase 6 |
| chr1_-_219450451 | 2.55 |
ENSRNOT00000025317
|
Rad9a
|
RAD9 checkpoint clamp component A |
| chr12_+_51878153 | 2.49 |
ENSRNOT00000056798
|
Hscb
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr15_-_11912806 | 2.43 |
ENSRNOT00000068171
ENSRNOT00000008759 ENSRNOT00000049771 |
Slc4a7
|
solute carrier family 4 member 7 |
| chr2_-_203043847 | 2.33 |
ENSRNOT00000067000
|
Man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr11_+_86890585 | 2.32 |
ENSRNOT00000002579
|
Ranbp1
|
RAN binding protein 1 |
| chr9_-_10441834 | 2.31 |
ENSRNOT00000043704
|
Rpl36
|
ribosomal protein L36 |
| chr3_-_148312791 | 2.30 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chr10_+_80790168 | 2.30 |
ENSRNOT00000073315
ENSRNOT00000075163 |
Car10
|
carbonic anhydrase 10 |
| chr10_+_89352835 | 2.28 |
ENSRNOT00000028060
|
Rpl27
|
ribosomal protein L27 |
| chr20_-_14282873 | 2.28 |
ENSRNOT00000001759
|
Adora2a
|
adenosine A2a receptor |
| chr9_+_42620006 | 2.28 |
ENSRNOT00000019966
|
Hs6st1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr9_-_46206605 | 2.26 |
ENSRNOT00000018640
|
Tbc1d8
|
TBC1 domain family, member 8 |
| chr1_+_260153645 | 2.25 |
ENSRNOT00000054717
|
Zfp518a
|
zinc finger protein 518A |
| chr1_+_88078350 | 2.25 |
ENSRNOT00000048677
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr7_-_2961873 | 2.22 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr12_+_660011 | 2.20 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr4_-_51844331 | 2.16 |
ENSRNOT00000003593
|
Gpr37
|
G protein-coupled receptor 37 |
| chr1_+_274310153 | 2.13 |
ENSRNOT00000054686
|
Smc3
|
structural maintenance of chromosomes 3 |
| chr3_+_80614937 | 2.12 |
ENSRNOT00000065462
|
Harbi1
|
harbinger transposase derived 1 |
| chr2_+_224851383 | 2.10 |
ENSRNOT00000015354
|
Alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr19_+_6046665 | 2.08 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chr20_-_48503898 | 2.06 |
ENSRNOT00000073091
|
Wasf1
|
WAS protein family, member 1 |
| chr1_+_165724451 | 2.03 |
ENSRNOT00000025827
|
Fam168a
|
family with sequence similarity 168, member A |
| chr3_+_147422095 | 2.03 |
ENSRNOT00000006786
|
Angpt4
|
angiopoietin 4 |
| chr5_+_128923934 | 2.02 |
ENSRNOT00000064145
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr11_+_31640407 | 2.01 |
ENSRNOT00000029985
ENSRNOT00000091472 |
Ifnar1
|
interferon alpha and beta receptor subunit 1 |
| chr13_+_36378356 | 1.98 |
ENSRNOT00000071305
|
C1ql2
|
complement C1q like 2 |
| chr2_+_30664639 | 1.98 |
ENSRNOT00000076372
ENSRNOT00000076294 ENSRNOT00000076434 ENSRNOT00000076484 |
Taf9
|
TATA-box binding protein associated factor 9 |
| chr13_+_110257571 | 1.96 |
ENSRNOT00000005715
|
Ints7
|
integrator complex subunit 7 |
| chr15_-_37663584 | 1.95 |
ENSRNOT00000012093
|
Cryl1
|
crystallin, lambda 1 |
| chr8_+_93439648 | 1.91 |
ENSRNOT00000043008
|
LOC100359563
|
ribosomal protein S20-like |
| chr13_+_21678512 | 1.89 |
ENSRNOT00000047108
|
Cntnap5b
|
contactin associated protein-like 5B |
| chr4_-_78342863 | 1.86 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr11_+_73198522 | 1.83 |
ENSRNOT00000002356
|
Xxylt1
|
xyloside xylosyltransferase 1 |
| chr6_-_95890325 | 1.82 |
ENSRNOT00000050217
|
LOC690335
|
similar to 60S ribosomal protein L23a |
| chr15_-_18493366 | 1.79 |
ENSRNOT00000010303
|
Kctd6
|
potassium channel tetramerization domain containing 6 |
| chr16_-_62239987 | 1.78 |
ENSRNOT00000020252
|
Gsr
|
glutathione-disulfide reductase |
| chr5_+_166870011 | 1.78 |
ENSRNOT00000088299
|
Rps2
|
ribosomal protein S2 |
| chr6_-_1622196 | 1.77 |
ENSRNOT00000007492
|
Prkd3
|
protein kinase D3 |
| chr16_+_20657099 | 1.77 |
ENSRNOT00000027053
|
Kxd1
|
KxDL motif containing 1 |
| chr7_+_117963740 | 1.73 |
ENSRNOT00000075405
|
LOC108348189
|
COMM domain-containing protein 5 |
| chrX_-_63203643 | 1.69 |
ENSRNOT00000065194
ENSRNOT00000076974 |
Zfx
|
zinc finger protein X-linked |
| chr8_-_67869019 | 1.65 |
ENSRNOT00000066009
|
Pias1
|
protein inhibitor of activated STAT, 1 |
| chr1_+_154377247 | 1.63 |
ENSRNOT00000092945
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_+_169115981 | 1.57 |
ENSRNOT00000067478
|
Olr135
|
olfactory receptor 135 |
| chr7_-_97902585 | 1.50 |
ENSRNOT00000008382
|
RGD1310852
|
similar to RIKEN cDNA 9130401M01 |
| chr8_+_69971778 | 1.42 |
ENSRNOT00000058007
ENSRNOT00000037941 ENSRNOT00000050649 |
Megf11
|
multiple EGF-like-domains 11 |
| chr6_-_27107030 | 1.40 |
ENSRNOT00000012558
|
Slc35f6
|
solute carrier family 35, member F6 |
| chr15_+_34270648 | 1.39 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr12_-_30106889 | 1.39 |
ENSRNOT00000065868
ENSRNOT00000001200 |
Tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr4_+_22081604 | 1.39 |
ENSRNOT00000067462
|
Crot
|
carnitine O-octanoyltransferase |
| chr8_+_22446661 | 1.35 |
ENSRNOT00000010030
|
Qtrt1
|
queuine tRNA-ribosyltransferase catalytic subunit 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 4.3 | 13.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 4.3 | 12.9 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) regulation of DNA replication origin binding(GO:1902595) |
| 3.6 | 14.3 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 3.4 | 13.6 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 2.9 | 8.8 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 2.7 | 18.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 2.6 | 10.5 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 2.6 | 13.0 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 2.6 | 12.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 2.2 | 11.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 2.1 | 8.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 2.1 | 6.4 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 1.9 | 17.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 1.9 | 5.6 | GO:0071049 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 1.8 | 11.0 | GO:0033227 | dsRNA transport(GO:0033227) |
| 1.8 | 10.8 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 1.7 | 13.8 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 1.7 | 5.0 | GO:1904700 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 1.5 | 4.5 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 1.4 | 12.9 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 1.4 | 6.9 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 1.3 | 10.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 1.3 | 5.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 1.3 | 3.8 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 1.3 | 3.8 | GO:0006272 | leading strand elongation(GO:0006272) |
| 1.3 | 6.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 1.2 | 6.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.2 | 3.7 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 1.2 | 7.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 1.2 | 2.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 1.1 | 3.4 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 1.1 | 6.6 | GO:1903377 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 1.0 | 4.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 1.0 | 6.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.0 | 4.0 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 1.0 | 14.0 | GO:0036065 | fucosylation(GO:0036065) |
| 1.0 | 3.9 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 1.0 | 11.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.9 | 4.7 | GO:0061741 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.9 | 7.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.8 | 3.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.8 | 2.3 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.8 | 2.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.7 | 8.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.7 | 3.7 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.7 | 12.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.7 | 2.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.7 | 6.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.6 | 4.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.6 | 5.6 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.6 | 6.5 | GO:2001140 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.6 | 4.7 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.6 | 2.9 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.6 | 2.3 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.6 | 20.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.6 | 3.3 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.5 | 6.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.5 | 1.6 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.5 | 13.8 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.5 | 3.7 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.5 | 2.6 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.5 | 4.6 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.5 | 1.4 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.4 | 4.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.4 | 3.0 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.4 | 3.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.4 | 2.1 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.4 | 3.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.4 | 4.8 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.4 | 4.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 2.9 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.4 | 2.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.4 | 1.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 1.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 2.9 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.3 | 1.8 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 2.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 4.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 2.3 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.3 | 5.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 2.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 2.0 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.3 | 1.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.3 | 3.9 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.3 | 4.1 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.3 | 4.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.3 | 3.8 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.3 | 2.2 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.3 | 3.8 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.2 | 2.7 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 6.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 10.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 1.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 5.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 | 1.3 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 4.6 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.2 | 7.7 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.2 | 0.8 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 | 6.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 0.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 1.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 1.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 4.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 3.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 2.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 3.5 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.2 | 1.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 2.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.2 | 2.0 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.2 | 0.9 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 1.8 | GO:0010041 | response to iron(III) ion(GO:0010041) |
| 0.1 | 5.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 5.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 8.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 1.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.9 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 6.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 2.8 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 5.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 5.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 3.7 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 6.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 2.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 10.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.9 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 3.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 3.1 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.1 | 1.7 | GO:0060746 | parental behavior(GO:0060746) |
| 0.1 | 6.7 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.1 | 2.0 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 5.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 6.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 1.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 2.3 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 1.3 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.7 | GO:0060707 | dorsal/ventral axis specification(GO:0009950) trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 5.7 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 3.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.9 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.3 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 1.9 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.3 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 2.1 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0000811 | GINS complex(GO:0000811) |
| 3.3 | 13.0 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 2.6 | 12.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 2.3 | 6.9 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 2.0 | 22.5 | GO:0042555 | MCM complex(GO:0042555) |
| 2.0 | 10.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.6 | 14.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 1.5 | 11.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.4 | 4.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.3 | 5.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.1 | 3.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 1.1 | 3.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 1.0 | 6.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.9 | 6.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.9 | 13.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.9 | 13.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.8 | 5.0 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.8 | 4.7 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.8 | 6.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 9.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.7 | 4.7 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.7 | 3.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.6 | 3.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.6 | 3.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.5 | 9.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.5 | 5.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.5 | 3.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.5 | 10.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.5 | 12.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 4.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 2.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.4 | 4.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 16.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 6.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 2.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 10.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 8.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.3 | 10.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 2.0 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.3 | 4.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 2.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 6.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 3.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.3 | 1.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 26.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 3.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 11.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 2.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 3.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 14.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 3.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 9.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 2.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 3.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 1.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 4.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 2.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 3.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 4.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 1.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 12.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 10.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 12.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 15.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 8.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 6.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.3 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.1 | 3.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 13.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 0.9 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 2.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 4.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 5.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.7 | GO:0000803 | sex chromosome(GO:0000803) XY body(GO:0001741) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 31.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 4.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 3.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 7.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 5.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.7 | GO:0005903 | brush border(GO:0005903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 14.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 2.2 | 11.0 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 2.2 | 6.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 2.1 | 8.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 2.1 | 6.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 1.9 | 9.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 1.7 | 21.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 1.6 | 4.7 | GO:0017098 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 1.5 | 6.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.4 | 4.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 1.3 | 3.9 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 1.3 | 14.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 1.3 | 3.9 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 1.3 | 3.9 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.2 | 3.7 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 1.0 | 5.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.0 | 6.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.0 | 18.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 1.0 | 2.9 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 1.0 | 3.8 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.9 | 14.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.9 | 4.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.9 | 7.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.9 | 13.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.9 | 14.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.8 | 4.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.8 | 10.6 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.8 | 23.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.8 | 14.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.7 | 2.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.6 | 16.7 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.6 | 4.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.6 | 2.9 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.5 | 3.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 4.5 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.5 | 1.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.5 | 1.4 | GO:0008458 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 0.5 | 2.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.4 | 5.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.4 | 1.3 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.4 | 3.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.4 | 2.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.4 | 2.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.4 | 11.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.3 | 2.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 1.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.3 | 6.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.3 | 4.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.3 | 4.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 3.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 4.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.3 | 14.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 1.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.3 | 4.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 1.8 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.3 | 13.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.3 | 1.8 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.3 | 2.3 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.2 | 2.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 2.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.2 | 10.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.2 | 1.9 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.2 | 1.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 6.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.2 | 0.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 3.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 2.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 3.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 5.6 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 1.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 3.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 5.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 5.0 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 2.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 5.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 4.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 3.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 12.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 8.8 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.1 | 2.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 7.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 12.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 2.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 7.5 | GO:0008144 | drug binding(GO:0008144) |
| 0.1 | 6.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 28.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 4.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 10.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 2.1 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 1.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.2 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 46.3 | PID ATM PATHWAY | ATM pathway |
| 0.4 | 24.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.4 | 6.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 23.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.3 | 4.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 5.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 4.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 6.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 4.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 8.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 1.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 8.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 7.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 2.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 2.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 11.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 31.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.4 | 28.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.3 | 14.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.8 | 12.8 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.8 | 13.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.6 | 8.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.5 | 9.7 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.5 | 4.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.5 | 14.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 5.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.5 | 13.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.4 | 4.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.4 | 5.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.4 | 6.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 6.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 8.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.3 | 28.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 10.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.3 | 10.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 7.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 1.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 11.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 6.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 3.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 6.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 6.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 3.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 2.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 4.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 2.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 4.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 4.4 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 2.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 5.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.0 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 4.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 2.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |