Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for E2f1
Z-value: 3.45
Transcription factors associated with E2f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f1
|
ENSRNOG00000016708 | E2F transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f1 | rn6_v1_chr3_-_150073721_150073721 | 0.73 | 1.1e-54 | Click! |
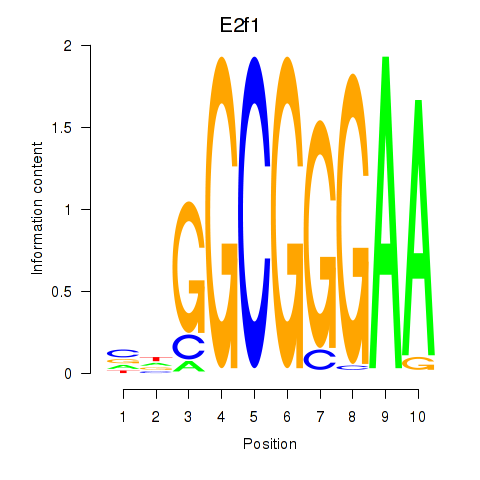
Activity profile of E2f1 motif
Sorted Z-values of E2f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_43617247 | 205.40 |
ENSRNOT00000075065
|
Hist1h2ao
|
histone cluster 1, H2ao |
| chr8_+_71514281 | 146.02 |
ENSRNOT00000022256
|
Ns5atp9
|
NS5A (hepatitis C virus) transactivated protein 9 |
| chr7_+_53275676 | 136.63 |
ENSRNOT00000031986
|
E2f7
|
E2F transcription factor 7 |
| chr13_-_110257367 | 119.58 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr3_+_159421671 | 102.26 |
ENSRNOT00000010343
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr3_+_59153280 | 91.43 |
ENSRNOT00000002066
|
Cdca7
|
cell division cycle associated 7 |
| chr9_-_26932201 | 90.77 |
ENSRNOT00000017081
|
Mcm3
|
minichromosome maintenance complex component 3 |
| chr8_-_66893083 | 88.23 |
ENSRNOT00000037028
ENSRNOT00000091755 |
Kif23
|
kinesin family member 23 |
| chr9_-_10757720 | 84.48 |
ENSRNOT00000083848
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr8_-_33661049 | 77.42 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_+_152995865 | 76.70 |
ENSRNOT00000014885
|
Il17ra
|
interleukin 17 receptor A |
| chr5_+_38844488 | 75.17 |
ENSRNOT00000009545
|
Mms22l
|
MMS22-like, DNA repair protein |
| chr18_+_51785111 | 74.02 |
ENSRNOT00000019351
|
Lmnb1
|
lamin B1 |
| chr9_-_10757954 | 73.34 |
ENSRNOT00000075265
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr7_-_3132457 | 72.74 |
ENSRNOT00000031963
|
Cdk2
|
cyclin dependent kinase 2 |
| chr10_-_85947938 | 71.67 |
ENSRNOT00000037318
ENSRNOT00000082427 |
Arl5c
|
ADP-ribosylation factor like GTPase 5C |
| chr7_+_2643288 | 71.28 |
ENSRNOT00000047241
|
Timeless
|
timeless circadian clock |
| chr1_-_94494980 | 70.55 |
ENSRNOT00000020014
|
Ccne1
|
cyclin E1 |
| chr4_-_122741110 | 69.58 |
ENSRNOT00000008888
|
Nup210
|
nucleoporin 210 |
| chr10_-_106828884 | 67.43 |
ENSRNOT00000071420
|
Tk1
|
thymidine kinase 1 |
| chr1_+_75356220 | 66.84 |
ENSRNOT00000019799
|
Lig1
|
DNA ligase 1 |
| chr13_-_113927745 | 66.74 |
ENSRNOT00000010910
ENSRNOT00000080616 |
Cr2
|
complement C3d receptor 2 |
| chr6_+_43884678 | 66.68 |
ENSRNOT00000091551
|
Rrm2
|
ribonucleotide reductase regulatory subunit M2 |
| chr4_-_77347011 | 65.86 |
ENSRNOT00000008149
|
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr20_-_3419831 | 65.51 |
ENSRNOT00000046798
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr15_-_44411004 | 65.21 |
ENSRNOT00000031163
|
Cdca2
|
cell division cycle associated 2 |
| chr5_+_144581427 | 63.30 |
ENSRNOT00000015227
|
Clspn
|
claspin |
| chr5_+_154522119 | 62.61 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr17_+_44520537 | 61.66 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr17_-_44815995 | 60.91 |
ENSRNOT00000091201
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr2_+_127686925 | 60.41 |
ENSRNOT00000086653
ENSRNOT00000016946 |
Plk4
|
polo-like kinase 4 |
| chr16_+_19800463 | 60.30 |
ENSRNOT00000023065
|
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr17_-_43627629 | 60.24 |
ENSRNOT00000022965
|
Hist1h2af
|
histone cluster 1, H2af |
| chr5_+_62718787 | 59.60 |
ENSRNOT00000011112
|
Galnt12
|
polypeptide N-acetylgalactosaminyltransferase 12 |
| chr6_+_91463308 | 59.50 |
ENSRNOT00000005595
|
Lrr1
|
leucine rich repeat protein 1 |
| chr11_-_66695353 | 58.75 |
ENSRNOT00000063995
|
Polq
|
DNA polymerase theta |
| chr2_-_30780121 | 57.52 |
ENSRNOT00000025129
|
Cenph
|
centromere protein H |
| chr9_+_81783349 | 56.14 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr1_+_242959488 | 55.46 |
ENSRNOT00000015668
|
Dock8
|
dedicator of cytokinesis 8 |
| chr1_-_167347662 | 54.83 |
ENSRNOT00000027641
ENSRNOT00000076592 |
Rhog
|
ras homolog family member G |
| chr6_+_144291974 | 54.69 |
ENSRNOT00000035255
|
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr5_+_59783890 | 54.21 |
ENSRNOT00000066277
|
Melk
|
maternal embryonic leucine zipper kinase |
| chr19_+_14523554 | 54.13 |
ENSRNOT00000084271
ENSRNOT00000064731 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr3_-_110021149 | 53.59 |
ENSRNOT00000007808
|
Fsip1
|
fibrous sheath interacting protein 1 |
| chr19_-_25220010 | 53.53 |
ENSRNOT00000008786
|
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr7_+_102586313 | 53.01 |
ENSRNOT00000006188
|
Myc
|
myelocytomatosis oncogene |
| chr1_+_167538263 | 51.64 |
ENSRNOT00000074058
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr1_+_82480195 | 51.08 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr6_-_28663233 | 50.72 |
ENSRNOT00000070979
|
Cenpo
|
centromere protein O |
| chr13_+_98924962 | 49.82 |
ENSRNOT00000074148
|
Lin9
|
lin-9 DREAM MuvB core complex component |
| chr5_-_163167299 | 49.53 |
ENSRNOT00000022478
|
Tnfrsf1b
|
TNF receptor superfamily member 1B |
| chr7_+_144078496 | 49.01 |
ENSRNOT00000055302
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr10_+_108132105 | 48.35 |
ENSRNOT00000072534
|
Cbx2
|
chromobox 2 |
| chr9_-_92963697 | 48.33 |
ENSRNOT00000023546
|
Gpr55
|
G protein-coupled receptor 55 |
| chr20_-_3397039 | 48.10 |
ENSRNOT00000001084
ENSRNOT00000085259 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr17_-_44520240 | 47.55 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr1_-_163129641 | 46.90 |
ENSRNOT00000083055
|
Capn5
|
calpain 5 |
| chr13_-_45068077 | 46.20 |
ENSRNOT00000004969
|
Mcm6
|
minichromosome maintenance complex component 6 |
| chr17_+_72429618 | 45.98 |
ENSRNOT00000026187
|
Gata3
|
GATA binding protein 3 |
| chr2_-_198412350 | 45.78 |
ENSRNOT00000040210
|
LOC100912489
|
histone H4-like |
| chr3_-_153250641 | 45.72 |
ENSRNOT00000008961
|
Samhd1
|
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
| chr2_-_207300854 | 45.25 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr7_+_27438351 | 45.25 |
ENSRNOT00000077976
|
AABR07056503.1
|
|
| chr20_-_6864571 | 44.69 |
ENSRNOT00000093253
|
Ppil1
|
peptidylprolyl isomerase like 1 |
| chr7_-_140617721 | 44.58 |
ENSRNOT00000081355
|
Tuba1b
|
tubulin, alpha 1B |
| chr17_+_44528125 | 44.08 |
ENSRNOT00000084538
|
LOC680322
|
similar to Histone H2A type 1 |
| chr15_-_24056073 | 44.08 |
ENSRNOT00000015100
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr9_+_112293388 | 43.80 |
ENSRNOT00000020767
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr10_-_4910305 | 43.56 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr11_-_31847490 | 43.19 |
ENSRNOT00000061304
|
Donson
|
downstream neighbor of SON |
| chr1_+_144831523 | 42.67 |
ENSRNOT00000039748
|
Mex3b
|
mex-3 RNA binding family member B |
| chr5_+_153507093 | 42.59 |
ENSRNOT00000086650
ENSRNOT00000083645 |
Runx3
|
runt-related transcription factor 3 |
| chr11_+_89511191 | 42.57 |
ENSRNOT00000002510
|
Mcm4
|
minichromosome maintenance complex component 4 |
| chr13_+_109713489 | 42.27 |
ENSRNOT00000004962
|
Batf3
|
basic leucine zipper ATF-like transcription factor 3 |
| chr5_-_129352886 | 42.22 |
ENSRNOT00000012088
|
Cdkn2c
|
cyclin-dependent kinase inhibitor 2C |
| chr1_+_257615088 | 41.42 |
ENSRNOT00000083183
|
LOC100911660
|
lymphocyte-specific helicase-like |
| chr7_+_37812831 | 41.31 |
ENSRNOT00000005910
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr7_-_31784192 | 41.13 |
ENSRNOT00000010869
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr7_-_119996824 | 41.08 |
ENSRNOT00000011079
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr14_-_3846891 | 41.04 |
ENSRNOT00000068520
|
Cdc7
|
cell division cycle 7 |
| chr7_+_120125633 | 40.93 |
ENSRNOT00000012480
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr4_-_170117325 | 40.83 |
ENSRNOT00000074198
|
LOC100912564
|
histone H4-like |
| chr20_+_20576377 | 40.69 |
ENSRNOT00000000783
ENSRNOT00000086806 |
Cdk1
|
cyclin-dependent kinase 1 |
| chr17_-_43770561 | 40.66 |
ENSRNOT00000088408
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr15_-_33193537 | 40.64 |
ENSRNOT00000016401
|
Haus4
|
HAUS augmin-like complex, subunit 4 |
| chrX_+_122507374 | 40.55 |
ENSRNOT00000032275
ENSRNOT00000080517 |
Dock11
|
dedicator of cytokinesis 11 |
| chr3_+_14299479 | 40.19 |
ENSRNOT00000092745
|
Cntrl
|
centriolin |
| chr8_+_48665652 | 40.11 |
ENSRNOT00000059715
|
H2afx
|
H2A histone family, member X |
| chr7_-_12741296 | 40.04 |
ENSRNOT00000060648
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr5_+_152721940 | 39.97 |
ENSRNOT00000039322
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr7_+_97984862 | 39.88 |
ENSRNOT00000008508
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr10_-_13619935 | 39.84 |
ENSRNOT00000064392
|
Ccnf
|
cyclin F |
| chr3_+_7422820 | 39.61 |
ENSRNOT00000064323
|
Ddx31
|
DEAD-box helicase 31 |
| chr16_+_62483761 | 38.58 |
ENSRNOT00000058805
|
Wrn
|
Werner syndrome RecQ like helicase |
| chr17_+_44522140 | 38.50 |
ENSRNOT00000080490
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr9_-_61528882 | 38.19 |
ENSRNOT00000015432
|
Ankrd44
|
ankyrin repeat domain 44 |
| chr19_-_914880 | 37.94 |
ENSRNOT00000017127
|
Cklf
|
chemokine-like factor |
| chr15_-_59215803 | 37.70 |
ENSRNOT00000032301
|
Lacc1
|
laccase domain containing 1 |
| chr16_+_48863418 | 37.55 |
ENSRNOT00000029868
|
Primpol
|
primase and DNA directed polymerase |
| chr5_-_76756140 | 37.47 |
ENSRNOT00000022107
ENSRNOT00000089251 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr6_-_7421456 | 37.46 |
ENSRNOT00000006725
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr1_+_199412834 | 37.37 |
ENSRNOT00000031891
|
Fus
|
fused in sarcoma RNA binding protein |
| chr10_-_19652898 | 37.29 |
ENSRNOT00000009648
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr10_-_108196217 | 37.23 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr12_+_1460538 | 37.16 |
ENSRNOT00000001444
|
Rfc3
|
replication factor C subunit 3 |
| chr4_+_161685258 | 37.13 |
ENSRNOT00000008012
ENSRNOT00000008003 |
Foxm1
|
forkhead box M1 |
| chr20_-_6864387 | 37.10 |
ENSRNOT00000068527
|
Ppil1
|
peptidylprolyl isomerase like 1 |
| chr16_-_19669153 | 36.93 |
ENSRNOT00000078714
ENSRNOT00000090775 |
Haus8
|
HAUS augmin-like complex, subunit 8 |
| chr8_-_39201588 | 36.91 |
ENSRNOT00000011226
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr6_-_11274932 | 36.75 |
ENSRNOT00000021538
|
Msh2
|
mutS homolog 2 |
| chr2_+_198359754 | 36.64 |
ENSRNOT00000048582
|
Hist2h2ab
|
histone cluster 2 H2A family member b |
| chr3_-_13435979 | 36.58 |
ENSRNOT00000029600
|
Pbx3
|
PBX homeobox 3 |
| chr5_-_152198813 | 36.52 |
ENSRNOT00000082953
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr1_-_154216340 | 36.15 |
ENSRNOT00000024082
|
Eed
|
embryonic ectoderm development |
| chr5_+_122647281 | 36.10 |
ENSRNOT00000066041
|
Mier1
|
mesoderm induction early response 1, transcriptional regulator |
| chr10_-_56511583 | 35.73 |
ENSRNOT00000021402
|
LOC497940
|
similar to RIKEN cDNA 2810408A11 |
| chr1_-_167347490 | 35.68 |
ENSRNOT00000076499
|
Rhog
|
ras homolog family member G |
| chr5_+_68717519 | 35.52 |
ENSRNOT00000066226
|
Smc2
|
structural maintenance of chromosomes 2 |
| chr1_-_225952516 | 35.38 |
ENSRNOT00000043387
|
Incenp
|
inner centromere protein |
| chr9_-_85445939 | 35.28 |
ENSRNOT00000072160
|
Ap1s3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr20_+_3155652 | 35.06 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr12_-_21720094 | 35.05 |
ENSRNOT00000032838
|
LOC100910636
|
zinc finger CW-type PWWP domain protein 1-like |
| chr20_-_3401273 | 34.96 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr12_+_24978483 | 34.88 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr10_-_70341837 | 34.74 |
ENSRNOT00000077261
|
Slfn13
|
schlafen family member 13 |
| chr1_+_85386470 | 34.64 |
ENSRNOT00000093332
ENSRNOT00000044326 |
Plekhg2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr3_+_148579920 | 34.53 |
ENSRNOT00000012432
|
Hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr7_-_12221330 | 34.45 |
ENSRNOT00000042204
|
Plk5
|
polo-like kinase 5 |
| chr10_+_11206226 | 34.05 |
ENSRNOT00000006979
|
Tfap4
|
transcription factor AP-4 |
| chr7_+_54213319 | 34.04 |
ENSRNOT00000005286
|
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr18_+_3162543 | 33.91 |
ENSRNOT00000078615
|
Rbbp8
|
RB binding protein 8, endonuclease |
| chr12_-_47588335 | 33.89 |
ENSRNOT00000001595
|
Ankrd13a
|
ankyrin repeat domain 13a |
| chr17_-_75886523 | 33.49 |
ENSRNOT00000046266
|
Usp6nl
|
USP6 N-terminal like |
| chr4_-_119742349 | 33.45 |
ENSRNOT00000013571
|
Isy1
|
ISY1 splicing factor homolog |
| chr10_+_45289741 | 33.42 |
ENSRNOT00000066190
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr5_+_24410863 | 33.36 |
ENSRNOT00000010591
|
Tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr10_+_17421075 | 33.31 |
ENSRNOT00000047011
|
Stk10
|
serine/threonine kinase 10 |
| chr1_-_142332588 | 33.26 |
ENSRNOT00000015065
|
Blm
|
Bloom syndrome RecQ like helicase |
| chr16_-_48921242 | 33.23 |
ENSRNOT00000041596
|
Cenpu
|
centromere protein U |
| chr8_-_78655856 | 33.17 |
ENSRNOT00000081185
|
Tcf12
|
transcription factor 12 |
| chr19_+_37652969 | 33.02 |
ENSRNOT00000041970
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr7_+_141882251 | 32.90 |
ENSRNOT00000087298
|
Atf1
|
activating transcription factor 1 |
| chr1_+_257614731 | 32.85 |
ENSRNOT00000075680
|
LOC100911660
|
lymphocyte-specific helicase-like |
| chr1_+_257766691 | 32.82 |
ENSRNOT00000088148
|
Hells
|
helicase, lymphoid specific |
| chr17_-_36177457 | 32.75 |
ENSRNOT00000036978
|
Mboat1
|
membrane bound O-acyltransferase domain containing 1 |
| chr1_-_136073483 | 32.73 |
ENSRNOT00000043854
ENSRNOT00000082564 ENSRNOT00000076340 |
Slco3a1
|
solute carrier organic anion transporter family, member 3a1 |
| chr7_-_12673659 | 32.49 |
ENSRNOT00000091650
ENSRNOT00000041277 ENSRNOT00000044865 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr16_+_60307148 | 32.27 |
ENSRNOT00000015239
|
Eri1
|
exoribonuclease 1 |
| chr1_-_32429917 | 32.27 |
ENSRNOT00000024111
|
Lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr12_-_46718355 | 32.06 |
ENSRNOT00000030031
|
Rab35
|
RAB35, member RAS oncogene family |
| chr19_-_39087880 | 32.03 |
ENSRNOT00000070822
|
Chtf8
|
chromosome transmission fidelity factor 8 |
| chr1_+_141120166 | 31.93 |
ENSRNOT00000050759
|
Fanci
|
Fanconi anemia, complementation group I |
| chr17_-_44527801 | 31.69 |
ENSRNOT00000089643
|
Hist1h2bk
|
histone cluster 1 H2B family member k |
| chr1_-_100554488 | 31.64 |
ENSRNOT00000026797
|
Pold1
|
DNA polymerase delta 1, catalytic subunit |
| chr2_+_189430041 | 31.45 |
ENSRNOT00000023567
ENSRNOT00000023605 |
Tpm3
|
tropomyosin 3 |
| chr5_-_77031120 | 31.44 |
ENSRNOT00000022987
|
Inip
|
INTS3 and NABP interacting protein |
| chr11_+_81358592 | 31.21 |
ENSRNOT00000002487
|
Rfc4
|
replication factor C subunit 4 |
| chr20_+_3156170 | 31.17 |
ENSRNOT00000082880
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr1_+_13212324 | 31.16 |
ENSRNOT00000089607
|
Reps1
|
RALBP1 associated Eps domain containing 1 |
| chr3_-_119619865 | 31.14 |
ENSRNOT00000016172
|
Itpripl1
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 1 |
| chr10_-_85049331 | 31.08 |
ENSRNOT00000012538
|
Tbx21
|
T-box 21 |
| chr12_-_31323810 | 30.88 |
ENSRNOT00000001247
|
Ran
|
RAN, member RAS oncogene family |
| chr5_+_150032999 | 30.68 |
ENSRNOT00000013301
|
Srsf4
|
serine and arginine rich splicing factor 4 |
| chr10_+_50928309 | 30.58 |
ENSRNOT00000033737
|
Hs3st3a1
|
heparan sulfate-glucosamine 3-sulfotransferase 3A1 |
| chr9_-_11027506 | 30.40 |
ENSRNOT00000071107
|
Chaf1a
|
chromatin assembly factor 1 subunit A |
| chr7_-_28711761 | 30.18 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr12_-_19314016 | 30.17 |
ENSRNOT00000001825
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr17_+_15814132 | 30.10 |
ENSRNOT00000032997
|
Susd3
|
sushi domain containing 3 |
| chr10_+_67427066 | 29.95 |
ENSRNOT00000035642
|
Atad5
|
ATPase family, AAA domain containing 5 |
| chrX_+_15098904 | 29.76 |
ENSRNOT00000007367
ENSRNOT00000087033 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr13_-_68858781 | 29.70 |
ENSRNOT00000065944
ENSRNOT00000092053 |
Rnf2
|
ring finger protein 2 |
| chr19_+_25120128 | 29.65 |
ENSRNOT00000081243
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr4_-_170145262 | 28.95 |
ENSRNOT00000074526
|
Hist1h4b
|
histone cluster 1, H4b |
| chr17_-_78812111 | 28.94 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr19_+_49457677 | 28.72 |
ENSRNOT00000084445
|
Cenpn
|
centromere protein N |
| chr17_+_89742140 | 28.55 |
ENSRNOT00000077899
|
Mastl
|
microtubule associated serine/threonine kinase-like |
| chr2_+_252018597 | 28.46 |
ENSRNOT00000020452
|
Mcoln2
|
mucolipin 2 |
| chr15_-_52443055 | 28.40 |
ENSRNOT00000087450
|
Xpo7
|
exportin 7 |
| chr1_+_167539036 | 28.32 |
ENSRNOT00000093112
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr11_+_34101197 | 28.31 |
ENSRNOT00000002299
|
Chaf1b
|
chromatin assembly factor 1 subunit B |
| chr12_-_24537313 | 28.31 |
ENSRNOT00000001975
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr18_-_27520295 | 28.11 |
ENSRNOT00000027544
|
Gfra3
|
GDNF family receptor alpha 3 |
| chr4_-_119889949 | 27.97 |
ENSRNOT00000033687
|
H1fx
|
H1 histone family, member X |
| chr4_-_159482869 | 27.78 |
ENSRNOT00000088333
|
Rad51ap1
|
RAD51 associated protein 1 |
| chr1_-_85408413 | 27.74 |
ENSRNOT00000093558
ENSRNOT00000026576 |
Rps16
|
ribosomal protein S16 |
| chr6_-_92643847 | 27.72 |
ENSRNOT00000009183
|
Pygl
|
glycogen phosphorylase L |
| chr17_+_17965881 | 27.49 |
ENSRNOT00000021751
|
Dek
|
DEK proto-oncogene |
| chr2_-_104133985 | 27.41 |
ENSRNOT00000088167
ENSRNOT00000067725 |
Pde7a
|
phosphodiesterase 7A |
| chr6_-_128149220 | 27.39 |
ENSRNOT00000014204
|
Gsc
|
goosecoid homeobox |
| chr1_-_221251329 | 27.35 |
ENSRNOT00000031678
|
Tigd3
|
tigger transposable element derived 3 |
| chr1_+_167538744 | 27.27 |
ENSRNOT00000093070
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr11_+_82373870 | 27.16 |
ENSRNOT00000002429
|
Tra2b
|
transformer 2 beta homolog (Drosophila) |
| chr3_+_72447969 | 27.14 |
ENSRNOT00000012022
|
Ssrp1
|
structure specific recognition protein 1 |
| chr1_-_84008293 | 27.03 |
ENSRNOT00000002053
|
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr3_+_37599728 | 26.85 |
ENSRNOT00000090575
|
Rif1
|
replication timing regulatory factor 1 |
| chr6_-_128727374 | 26.66 |
ENSRNOT00000082152
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr6_+_126636491 | 26.54 |
ENSRNOT00000089601
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr1_+_142034423 | 26.44 |
ENSRNOT00000090006
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr16_+_19669411 | 26.41 |
ENSRNOT00000086105
ENSRNOT00000087576 ENSRNOT00000083651 ENSRNOT00000045099 |
Myo9b
|
myosin IXb |
| chr4_+_71740532 | 26.18 |
ENSRNOT00000023537
|
Zyx
|
zyxin |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 29.4 | 88.2 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 27.6 | 220.8 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 27.3 | 136.6 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 27.1 | 461.0 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 24.6 | 73.7 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 24.5 | 122.7 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 24.2 | 72.7 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 23.8 | 71.3 | GO:1904976 | cellular response to bleomycin(GO:1904976) |
| 23.4 | 117.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 22.4 | 67.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 22.0 | 87.8 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 17.7 | 53.0 | GO:0072185 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) |
| 17.0 | 118.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 16.9 | 67.4 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) |
| 16.7 | 66.7 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 16.1 | 145.3 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 15.1 | 211.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 14.7 | 58.7 | GO:0097681 | double-strand break repair via alternative nonhomologous end joining(GO:0097681) |
| 14.0 | 84.3 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 13.4 | 40.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 13.1 | 209.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 12.5 | 37.6 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 12.5 | 312.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 12.0 | 60.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 11.6 | 34.9 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 11.6 | 34.7 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 11.5 | 46.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 11.4 | 45.7 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 11.3 | 78.9 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 11.2 | 56.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 11.1 | 33.4 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 11.1 | 33.4 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 11.0 | 65.9 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 10.8 | 32.5 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 10.8 | 32.3 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 10.7 | 10.7 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 10.0 | 39.8 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 9.5 | 66.5 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 9.4 | 46.9 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 9.4 | 65.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 9.4 | 37.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 9.4 | 28.1 | GO:1905073 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 9.2 | 27.5 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 9.1 | 81.6 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 9.1 | 81.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 9.0 | 63.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 9.0 | 26.9 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 8.7 | 26.2 | GO:0070637 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 8.5 | 85.4 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 8.5 | 42.7 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 8.4 | 25.3 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 8.4 | 25.1 | GO:2000111 | senescence-associated heterochromatin focus assembly(GO:0035986) positive regulation of macrophage apoptotic process(GO:2000111) |
| 8.4 | 16.7 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 8.3 | 33.0 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 8.2 | 131.2 | GO:0051383 | kinetochore organization(GO:0051383) |
| 8.0 | 24.0 | GO:2001226 | negative regulation of inorganic anion transmembrane transport(GO:1903796) negative regulation of chloride transport(GO:2001226) |
| 7.9 | 23.7 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 7.9 | 47.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 7.7 | 15.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 7.6 | 15.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 7.5 | 29.9 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 7.5 | 22.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 7.3 | 44.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 7.0 | 27.9 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 6.9 | 20.8 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 6.9 | 27.4 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 6.7 | 20.2 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 6.7 | 20.1 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 6.7 | 33.4 | GO:0042148 | strand invasion(GO:0042148) |
| 6.6 | 19.7 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 6.6 | 45.9 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 6.5 | 26.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 6.5 | 19.4 | GO:1902961 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 6.4 | 19.3 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 6.4 | 25.6 | GO:2001206 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) TNFSF11-mediated signaling pathway(GO:0071847) positive regulation of osteoclast development(GO:2001206) |
| 6.3 | 31.6 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 6.3 | 18.9 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 6.3 | 18.8 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 6.2 | 68.0 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 6.1 | 79.4 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 6.1 | 61.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 6.1 | 18.3 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 6.0 | 24.0 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 5.7 | 17.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 5.7 | 33.9 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 5.6 | 16.8 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 5.6 | 11.2 | GO:0099542 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 5.4 | 59.8 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 5.4 | 48.3 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 5.4 | 32.1 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 5.3 | 15.8 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 5.3 | 36.9 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 5.2 | 47.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 5.2 | 20.8 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 5.2 | 41.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 5.1 | 30.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 5.1 | 25.6 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 5.1 | 10.2 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 5.1 | 15.2 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 5.1 | 35.5 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 5.0 | 20.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 5.0 | 19.8 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 4.9 | 14.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 4.9 | 68.9 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 4.9 | 19.5 | GO:1900623 | regulation of mast cell cytokine production(GO:0032763) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 4.9 | 19.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 4.8 | 33.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 4.8 | 124.8 | GO:0031297 | replication fork processing(GO:0031297) |
| 4.8 | 23.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 4.7 | 33.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 4.7 | 18.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 4.6 | 32.3 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 4.6 | 13.8 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 4.6 | 9.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 4.6 | 13.7 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 4.5 | 9.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 4.5 | 36.1 | GO:0061085 | regulation of histone H3-K27 methylation(GO:0061085) |
| 4.5 | 9.0 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 4.5 | 40.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 4.3 | 21.7 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 4.3 | 25.9 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 4.3 | 12.9 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 4.3 | 29.9 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 4.2 | 50.9 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 4.2 | 12.6 | GO:1903969 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) regulation of prolactin signaling pathway(GO:1902211) regulation of interleukin-4-mediated signaling pathway(GO:1902214) regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 4.2 | 16.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 4.2 | 37.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 4.1 | 12.4 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 4.1 | 282.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 4.0 | 15.8 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 3.9 | 11.7 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 3.9 | 15.7 | GO:0042891 | antibiotic transport(GO:0042891) |
| 3.9 | 11.6 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 3.8 | 15.3 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 3.8 | 19.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 3.7 | 22.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 3.7 | 14.9 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 3.7 | 29.6 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 3.7 | 29.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 3.6 | 10.8 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 3.6 | 17.9 | GO:1901977 | negative regulation of cell cycle checkpoint(GO:1901977) negative regulation of DNA damage checkpoint(GO:2000002) |
| 3.5 | 21.0 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 3.5 | 14.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 3.5 | 24.2 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 3.4 | 6.7 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 3.4 | 13.4 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 3.3 | 26.7 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 3.3 | 55.5 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 3.2 | 9.7 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 3.2 | 22.6 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 3.2 | 9.7 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 3.2 | 16.0 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 3.2 | 9.6 | GO:1903699 | tarsal gland development(GO:1903699) |
| 3.2 | 15.9 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 3.2 | 12.7 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 3.1 | 15.7 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 3.1 | 181.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 3.1 | 90.5 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 3.1 | 49.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 3.1 | 30.6 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 3.0 | 57.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 3.0 | 23.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 3.0 | 17.7 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 3.0 | 11.8 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 2.9 | 11.7 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 2.9 | 20.3 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 2.9 | 8.6 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 2.8 | 42.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 2.8 | 22.7 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 2.8 | 11.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 2.8 | 11.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 2.8 | 13.8 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 2.7 | 16.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 2.7 | 24.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 2.7 | 21.4 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 2.7 | 8.0 | GO:2000342 | positive regulation of hemoglobin biosynthetic process(GO:0046985) negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 2.7 | 8.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 2.6 | 26.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 2.6 | 18.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 2.5 | 14.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 2.5 | 12.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 2.4 | 26.4 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 2.4 | 14.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 2.4 | 2.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 2.4 | 33.1 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 2.4 | 11.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 2.4 | 16.5 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 2.3 | 71.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 2.3 | 6.8 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 2.3 | 6.8 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 2.2 | 24.5 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 2.2 | 74.8 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 2.2 | 193.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 2.2 | 36.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 2.1 | 51.0 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 2.1 | 8.5 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 2.1 | 42.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 2.1 | 6.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 2.1 | 14.7 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 2.1 | 94.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 2.1 | 27.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 2.1 | 16.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 2.1 | 10.4 | GO:0032202 | telomere assembly(GO:0032202) |
| 2.1 | 12.4 | GO:0046477 | galactosylceramide metabolic process(GO:0006681) glycosylceramide catabolic process(GO:0046477) |
| 2.1 | 31.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 2.1 | 8.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 2.1 | 10.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 2.1 | 32.9 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 2.0 | 36.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 2.0 | 18.3 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 2.0 | 20.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 2.0 | 11.9 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 2.0 | 13.9 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 2.0 | 13.7 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 1.9 | 13.6 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 1.9 | 13.6 | GO:0015871 | choline transport(GO:0015871) |
| 1.9 | 20.6 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 1.9 | 11.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 1.8 | 1.8 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 1.8 | 9.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 1.8 | 42.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 1.8 | 34.5 | GO:0002347 | response to tumor cell(GO:0002347) |
| 1.8 | 7.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 1.8 | 32.4 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 1.8 | 21.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 1.7 | 12.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 1.7 | 67.7 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 1.7 | 49.0 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 1.7 | 5.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 1.6 | 13.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 1.6 | 4.9 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 1.6 | 11.2 | GO:0035878 | nail development(GO:0035878) |
| 1.6 | 14.3 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 1.5 | 10.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 1.5 | 10.7 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 1.5 | 9.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 1.5 | 7.6 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 1.5 | 4.6 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 1.5 | 27.4 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 1.5 | 4.5 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 1.5 | 4.4 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 1.5 | 13.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 1.5 | 5.8 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 1.4 | 33.3 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 1.4 | 13.0 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 1.4 | 2.9 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 1.4 | 2.8 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 1.4 | 11.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 1.4 | 5.5 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 1.4 | 9.7 | GO:0015705 | iodide transport(GO:0015705) |
| 1.4 | 37.1 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 1.4 | 17.6 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 1.4 | 9.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.3 | 10.6 | GO:0080009 | mRNA methylation(GO:0080009) |
| 1.3 | 26.4 | GO:0070266 | necroptotic process(GO:0070266) |
| 1.3 | 11.8 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 1.3 | 51.0 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 1.3 | 2.5 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 1.3 | 3.8 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 1.3 | 48.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 1.2 | 7.5 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 1.2 | 43.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 1.2 | 83.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 1.2 | 14.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 1.2 | 28.1 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 1.2 | 13.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 1.2 | 6.1 | GO:0060148 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 1.2 | 46.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 1.2 | 4.8 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 1.2 | 25.0 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 1.2 | 9.4 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 1.2 | 4.7 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 1.2 | 3.5 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 1.2 | 7.0 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 1.2 | 4.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 1.1 | 3.4 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 1.1 | 8.0 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 1.1 | 5.7 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 1.1 | 3.4 | GO:0009751 | response to salicylic acid(GO:0009751) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 1.1 | 6.8 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 1.1 | 4.5 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 1.1 | 12.3 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 1.1 | 11.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 1.1 | 13.3 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 1.1 | 4.3 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 1.1 | 3.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.1 | 2.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 1.1 | 4.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 1.1 | 10.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.1 | 66.7 | GO:0042100 | B cell proliferation(GO:0042100) |
| 1.0 | 7.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 1.0 | 2.1 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 1.0 | 12.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 1.0 | 20.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 1.0 | 14.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 1.0 | 15.3 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 1.0 | 21.4 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 1.0 | 3.0 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 1.0 | 22.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 1.0 | 17.3 | GO:0030903 | notochord development(GO:0030903) |
| 1.0 | 99.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 1.0 | 46.9 | GO:0007051 | spindle organization(GO:0007051) |
| 0.9 | 6.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.9 | 7.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.9 | 13.5 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.9 | 7.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.9 | 7.0 | GO:0033260 | nuclear DNA replication(GO:0033260) |
| 0.9 | 3.5 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.9 | 15.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.9 | 2.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.9 | 31.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.8 | 4.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.8 | 5.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.8 | 22.0 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.8 | 8.0 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.8 | 36.8 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.8 | 8.6 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.8 | 31.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.7 | 6.7 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.7 | 18.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.7 | 5.2 | GO:0097240 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.7 | 136.2 | GO:0006281 | DNA repair(GO:0006281) |
| 0.7 | 11.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.7 | 5.6 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.7 | 8.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.7 | 8.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.7 | 178.7 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.6 | 13.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.6 | 3.8 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.6 | 14.0 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.6 | 7.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.6 | 27.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.6 | 8.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.6 | 2.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.6 | 7.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.6 | 5.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.6 | 17.1 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.5 | 13.2 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.5 | 20.5 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.5 | 2.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.5 | 1.0 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.5 | 7.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.5 | 2.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.5 | 12.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.5 | 11.3 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.5 | 3.2 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.4 | 3.9 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.4 | 11.7 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.4 | 3.3 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.4 | 7.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.4 | 4.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.4 | 5.8 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.4 | 1.2 | GO:0048625 | branchiomeric skeletal muscle development(GO:0014707) myoblast fate commitment(GO:0048625) |
| 0.4 | 6.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.4 | 9.7 | GO:0007567 | parturition(GO:0007567) |
| 0.4 | 9.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.4 | 2.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.4 | 3.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 14.9 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.4 | 14.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.4 | 5.2 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.4 | 17.7 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.4 | 14.4 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.3 | 1.4 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.3 | 8.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.3 | 7.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.3 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.3 | 12.1 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.3 | 1.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.3 | 12.4 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.3 | 8.6 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.3 | 2.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.3 | 15.7 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.3 | 0.6 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.3 | 0.9 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.3 | 7.5 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.3 | 3.9 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.3 | 2.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 14.9 | GO:0042113 | B cell activation(GO:0042113) |
| 0.2 | 1.5 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 8.6 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.2 | 1.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 6.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 3.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 2.5 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.2 | 1.7 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.2 | 3.7 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.2 | 0.4 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 1.8 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.2 | 5.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 2.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 2.8 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.1 | 2.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.4 | GO:0048333 | mesodermal cell differentiation(GO:0048333) |
| 0.1 | 7.2 | GO:0048144 | fibroblast proliferation(GO:0048144) |
| 0.1 | 1.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 1.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 7.6 | GO:0071222 | cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 0.6 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 14.4 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 1.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 15.9 | GO:0045137 | development of primary sexual characteristics(GO:0045137) |
| 0.1 | 4.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 3.7 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0061377 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 43.5 | 173.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 32.9 | 361.7 | GO:0042555 | MCM complex(GO:0042555) |
| 24.2 | 72.7 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 23.7 | 118.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 22.1 | 88.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 20.5 | 123.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 19.6 | 58.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 18.1 | 54.4 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 17.1 | 480.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 13.6 | 40.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 12.1 | 48.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 12.1 | 60.4 | GO:0098536 | deuterosome(GO:0098536) |
| 11.6 | 92.6 | GO:0000796 | condensin complex(GO:0000796) |
| 11.2 | 78.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 11.0 | 66.0 | GO:0001740 | Barr body(GO:0001740) |
| 11.0 | 32.9 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 10.2 | 51.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 9.4 | 85.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 8.9 | 88.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 8.6 | 68.8 | GO:0005638 | lamin filament(GO:0005638) |
| 8.6 | 85.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 8.2 | 41.1 | GO:0043293 | apoptosome(GO:0043293) |
| 8.0 | 32.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 8.0 | 55.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 7.7 | 30.9 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 7.7 | 38.6 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 7.4 | 66.5 | GO:0005688 | U6 snRNP(GO:0005688) |
| 7.0 | 28.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 7.0 | 34.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 7.0 | 27.9 | GO:0018444 | translation release factor complex(GO:0018444) |
| 6.8 | 149.5 | GO:0045120 | pronucleus(GO:0045120) |
| 6.7 | 33.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 6.6 | 32.8 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 6.5 | 274.8 | GO:0000791 | euchromatin(GO:0000791) |
| 6.4 | 19.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 6.4 | 50.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 6.3 | 31.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 6.3 | 25.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 6.2 | 43.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 5.7 | 355.8 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 5.4 | 16.3 | GO:0000811 | GINS complex(GO:0000811) |
| 5.1 | 35.5 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 5.0 | 19.8 | GO:0071914 | prominosome(GO:0071914) |
| 4.8 | 14.4 | GO:0044609 | DBIRD complex(GO:0044609) |
| 4.8 | 19.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 4.7 | 46.8 | GO:0070187 | telosome(GO:0070187) |
| 4.6 | 18.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 4.5 | 13.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 4.5 | 31.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 4.5 | 22.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 4.5 | 93.9 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 4.4 | 21.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 4.3 | 52.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 4.3 | 17.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 4.3 | 29.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 4.3 | 42.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 4.2 | 67.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 4.2 | 66.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 4.1 | 40.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 3.8 | 26.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 3.8 | 41.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 3.7 | 96.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 3.6 | 32.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 3.4 | 40.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 3.4 | 47.5 | GO:0001741 | XY body(GO:0001741) |
| 3.2 | 22.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 3.2 | 34.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 3.2 | 22.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 3.1 | 27.9 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 2.9 | 8.6 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 2.9 | 8.6 | GO:0005745 | m-AAA complex(GO:0005745) |
| 2.8 | 19.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 2.8 | 11.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 2.7 | 26.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 2.6 | 15.8 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 2.5 | 29.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 2.5 | 49.5 | GO:0043196 | varicosity(GO:0043196) |
| 2.5 | 7.4 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 2.4 | 14.5 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 2.4 | 9.6 | GO:0032021 | NELF complex(GO:0032021) |
| 2.3 | 27.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 2.3 | 78.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 2.2 | 11.2 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 2.2 | 13.3 | GO:0090543 | annulate lamellae(GO:0005642) Flemming body(GO:0090543) |
| 2.2 | 93.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 2.1 | 8.4 | GO:0072487 | MSL complex(GO:0072487) |
| 2.1 | 18.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 2.0 | 122.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 1.9 | 174.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 1.9 | 15.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 1.8 | 32.0 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 1.8 | 19.4 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 1.8 | 21.1 | GO:0016342 | catenin complex(GO:0016342) |
| 1.7 | 12.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 1.7 | 8.6 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.7 | 6.8 | GO:0000125 | PCAF complex(GO:0000125) |
| 1.7 | 16.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 1.7 | 191.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 1.7 | 150.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 1.6 | 16.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 1.6 | 24.3 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 1.6 | 12.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.6 | 9.4 | GO:0036396 | MIS complex(GO:0036396) |
| 1.5 | 1.5 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 1.4 | 22.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.4 | 13.8 | GO:0002177 | manchette(GO:0002177) |
| 1.4 | 6.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 1.3 | 24.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 1.3 | 13.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.3 | 430.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 1.3 | 13.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 1.3 | 12.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 1.3 | 10.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.2 | 7.5 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 1.2 | 112.3 | GO:0005814 | centriole(GO:0005814) |
| 1.1 | 10.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 1.1 | 539.6 | GO:0005694 | chromosome(GO:0005694) |
| 1.1 | 19.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 1.1 | 3.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 1.0 | 34.9 | GO:0016235 | aggresome(GO:0016235) |
| 1.0 | 76.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 1.0 | 11.2 | GO:0097526 | spliceosomal tri-snRNP complex(GO:0097526) |
| 1.0 | 15.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.0 | 8.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.9 | 29.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.8 | 16.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.8 | 6.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.8 | 8.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.8 | 28.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.8 | 49.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.8 | 21.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.7 | 30.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.7 | 29.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.7 | 47.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.7 | 2.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.7 | 4.9 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.7 | 23.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.7 | 7.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.6 | 2.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 33.4 | GO:0016605 | PML body(GO:0016605) |
| 0.6 | 47.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.6 | 15.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.6 | 50.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.6 | 49.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.6 | 24.0 | GO:0030286 | dynein complex(GO:0030286) |
| 0.6 | 2.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.5 | 12.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.5 | 12.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.5 | 21.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.5 | 9.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.5 | 21.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.5 | 25.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.5 | 1.4 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.5 | 173.6 | GO:0016604 | nuclear body(GO:0016604) |
| 0.5 | 3.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.4 | 19.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.4 | 1.2 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.4 | 5.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.4 | 489.6 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.3 | 8.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 18.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.3 | 2.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.3 | 1.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 7.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.3 | 3.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 5.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 0.8 | GO:1903349 | omegasome membrane(GO:1903349) omegasome(GO:1990462) |
| 0.3 | 1.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.3 | 2.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 3.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 7.6 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 3.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 2.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 24.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 3.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 2.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 9.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 37.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 9.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 10.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 6.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 32.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.9 | GO:0070993 | eukaryotic 43S preinitiation complex(GO:0016282) translation preinitiation complex(GO:0070993) |
| 0.1 | 3.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.5 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 2.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 39.5 | 157.8 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 29.0 | 173.9 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 18.0 | 71.8 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 17.3 | 51.9 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 16.9 | 67.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 15.5 | 155.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 12.9 | 64.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 12.6 | 88.4 | GO:0003896 | DNA primase activity(GO:0003896) |
| 12.4 | 37.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 12.1 | 48.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 11.6 | 34.9 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 11.2 | 11.2 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 10.8 | 32.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 10.8 | 107.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 10.5 | 94.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 10.2 | 51.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 9.8 | 49.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 9.8 | 58.7 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 9.6 | 76.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 9.2 | 46.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 9.1 | 63.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 9.0 | 36.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 9.0 | 17.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 8.6 | 25.8 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 8.4 | 16.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 8.1 | 32.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 7.7 | 30.9 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 7.6 | 68.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 7.5 | 75.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 7.4 | 29.7 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 7.2 | 35.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 6.8 | 27.0 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 6.7 | 33.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 6.6 | 290.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 6.5 | 26.2 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 6.5 | 64.9 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 6.4 | 89.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 6.3 | 25.4 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 6.3 | 18.8 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 6.1 | 18.3 | GO:0070976 | TIR domain binding(GO:0070976) |
| 6.1 | 66.7 | GO:0004875 | complement receptor activity(GO:0004875) |
| 5.9 | 17.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 5.6 | 16.7 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 5.5 | 158.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 5.5 | 21.9 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 5.4 | 43.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 5.2 | 15.7 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 4.9 | 97.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 4.8 | 33.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 4.8 | 19.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 4.7 | 28.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 4.7 | 28.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 4.6 | 13.9 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 4.5 | 31.6 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 4.5 | 31.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 4.3 | 21.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 4.3 | 12.9 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 4.3 | 845.7 | GO:0042393 | histone binding(GO:0042393) |
| 4.2 | 29.6 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) |
| 4.2 | 21.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 4.2 | 12.6 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 4.1 | 82.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 4.1 | 24.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 4.1 | 4.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 4.0 | 44.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 4.0 | 64.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 3.9 | 11.8 | GO:0009041 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 3.9 | 50.6 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 3.8 | 49.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 3.8 | 26.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 3.7 | 100.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 3.7 | 22.2 | GO:0046977 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 3.6 | 32.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 3.5 | 42.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 3.5 | 24.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 3.5 | 20.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 3.4 | 13.8 | GO:0035877 | death effector domain binding(GO:0035877) |
| 3.4 | 10.3 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 3.3 | 43.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 3.3 | 16.5 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 3.2 | 18.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 3.1 | 58.8 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 3.1 | 21.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 3.1 | 30.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 2.9 | 8.8 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 2.9 | 17.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 2.9 | 11.6 | GO:0019002 | GMP binding(GO:0019002) |
| 2.8 | 33.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 2.8 | 11.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 2.7 | 98.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 2.7 | 27.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 2.7 | 16.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 2.7 | 10.7 | GO:0001156 | transcription factor activity, core RNA polymerase III binding(GO:0000995) transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) TFIIIC-class transcription factor binding(GO:0001156) |
| 2.6 | 7.8 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 2.6 | 15.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 2.6 | 113.4 | GO:0070888 | E-box binding(GO:0070888) |
| 2.6 | 10.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 2.6 | 17.9 | GO:0017070 | U6 snRNA binding(GO:0017070) U4 snRNA binding(GO:0030621) |
| 2.5 | 20.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 2.5 | 27.9 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 2.5 | 49.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 2.5 | 9.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 2.5 | 22.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 2.5 | 24.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 2.4 | 17.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 2.3 | 6.8 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 2.2 | 13.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 2.2 | 45.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 2.1 | 21.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 2.1 | 19.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 2.1 | 54.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 2.1 | 39.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 2.1 | 22.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 2.1 | 82.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 2.0 | 34.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 2.0 | 38.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 2.0 | 14.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 2.0 | 25.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 2.0 | 17.7 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 1.9 | 19.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 1.9 | 22.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.9 | 16.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 1.9 | 72.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 1.9 | 16.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 1.9 | 53.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 1.8 | 9.2 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 1.8 | 8.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 1.7 | 14.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 1.7 | 11.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.7 | 10.2 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.7 | 102.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 1.7 | 34.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 1.7 | 113.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 1.6 | 4.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 1.6 | 32.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 1.6 | 3.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 1.6 | 38.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 1.6 | 9.6 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 1.6 | 15.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 1.5 | 27.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 1.5 | 4.5 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 1.5 | 11.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.5 | 16.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 1.4 | 57.8 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 1.4 | 4.2 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 1.4 | 37.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 1.4 | 37.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 1.4 | 71.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 1.4 | 4.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 1.4 | 19.0 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.4 | 6.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 1.3 | 30.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 1.3 | 10.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.3 | 10.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 1.3 | 27.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 1.3 | 78.7 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 1.3 | 179.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 1.3 | 2.5 | GO:0005119 | smoothened binding(GO:0005119) |
| 1.2 | 6.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 1.2 | 12.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 1.2 | 8.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 1.2 | 21.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 1.2 | 9.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 1.2 | 9.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 1.2 | 39.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 1.2 | 10.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 1.1 | 3.3 | GO:0033222 | xylose binding(GO:0033222) |
| 1.1 | 21.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 1.1 | 3.3 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 1.0 | 37.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 1.0 | 22.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 1.0 | 35.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 1.0 | 10.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.0 | 14.8 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 1.0 | 14.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 1.0 | 24.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 1.0 | 17.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 1.0 | 7.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.9 | 10.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.9 | 5.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.9 | 73.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.9 | 7.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.9 | 31.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.8 | 4.2 | GO:0070990 | snRNP binding(GO:0070990) U1 snRNP binding(GO:1990446) |
| 0.8 | 9.2 | GO:0005346 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.8 | 3.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.8 | 7.5 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.8 | 5.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.8 | 43.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.8 | 8.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.8 | 8.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.8 | 13.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.8 | 30.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.8 | 6.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.7 | 15.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.7 | 3.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.7 | 5.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.7 | 24.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.7 | 14.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.7 | 9.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.7 | 30.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.7 | 185.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.7 | 15.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.6 | 9.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.6 | 13.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.6 | 40.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.6 | 4.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.6 | 11.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.6 | 13.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.6 | 17.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.6 | 15.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.6 | 9.1 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.6 | 7.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.5 | 713.2 | GO:0003677 | DNA binding(GO:0003677) |
| 0.5 | 4.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 3.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.5 | 44.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.5 | 8.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.5 | 13.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.5 | 17.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.5 | 16.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.5 | 3.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.5 | 13.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.4 | 1.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.4 | 15.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.4 | 10.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.4 | 3.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 1.9 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.4 | 4.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.4 | 6.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 13.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.3 | 3.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.3 | 11.4 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.3 | 3.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 13.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 4.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.3 | 23.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.3 | 7.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.3 | 7.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 2.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 3.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 15.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 8.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.3 | 5.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.3 | 23.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.3 | 43.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.3 | 34.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.3 | 6.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.5 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.3 | 6.8 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.2 | 8.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 5.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 4.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 1.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 2.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 45.3 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.2 | 14.6 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.2 | 4.0 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 0.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 12.4 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 0.9 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 7.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 42.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 1.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 3.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 7.4 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 3.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 5.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 18.9 | GO:0003723 | RNA binding(GO:0003723) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.9 | 253.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 11.4 | 843.0 | PID E2F PATHWAY | E2F transcription factor network |
| 7.5 | 309.4 | PID ATR PATHWAY | ATR signaling pathway |
| 6.9 | 201.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 5.6 | 100.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 4.1 | 24.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 3.8 | 99.0 | PID ATM PATHWAY | ATM pathway |
| 3.7 | 11.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 3.5 | 159.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 3.4 | 34.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 3.3 | 9.9 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 3.1 | 49.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 3.0 | 77.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 2.7 | 21.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 2.4 | 26.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 2.3 | 49.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 2.2 | 21.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 2.2 | 107.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 2.0 | 15.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 1.9 | 37.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 1.9 | 67.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 1.8 | 52.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 1.8 | 44.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 1.7 | 14.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 1.6 | 82.1 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 1.6 | 14.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 1.5 | 25.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 1.5 | 156.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 1.5 | 68.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 1.4 | 31.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 1.4 | 102.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 1.4 | 17.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 1.3 | 61.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 1.1 | 13.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 1.1 | 17.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 1.1 | 41.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 1.0 | 23.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.9 | 16.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.9 | 30.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.8 | 60.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.8 | 28.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.7 | 37.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.7 | 26.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.7 | 21.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.7 | 13.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.7 | 16.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.7 | 24.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.7 | 10.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.7 | 10.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.6 | 25.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.6 | 15.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.6 | 21.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.6 | 6.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.5 | 21.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.5 | 2.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 7.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.4 | 5.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.3 | 12.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 2.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.2 | 2.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 6.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 25.3 | 278.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 13.4 | 320.7 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 12.5 | 337.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 9.7 | 68.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 8.2 | 41.0 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 8.2 | 147.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 8.1 | 32.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 6.8 | 170.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 6.6 | 125.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 6.3 | 144.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 6.1 | 61.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 4.8 | 24.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 4.7 | 102.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 4.4 | 128.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 4.4 | 61.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 4.3 | 43.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 4.3 | 34.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 4.3 | 21.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 4.1 | 77.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 3.9 | 11.7 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 3.7 | 18.4 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 3.7 | 117.3 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 3.4 | 47.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 3.3 | 271.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 3.2 | 32.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 3.2 | 59.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 3.0 | 81.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 2.9 | 63.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 2.9 | 40.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 2.7 | 52.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 2.7 | 67.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 2.6 | 15.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 2.5 | 86.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 2.5 | 44.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 2.3 | 13.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 2.3 | 48.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 2.2 | 11.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 2.2 | 71.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 2.2 | 55.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 2.1 | 12.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 2.1 | 21.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 2.0 | 32.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 2.0 | 25.5 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 1.9 | 33.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 1.8 | 45.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 1.8 | 21.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 1.7 | 32.9 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 1.7 | 52.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.7 | 11.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 1.6 | 16.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.6 | 16.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 1.6 | 11.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 1.6 | 45.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 1.5 | 21.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 1.4 | 15.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 1.4 | 25.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 1.4 | 16.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 1.4 | 62.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 1.3 | 12.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 1.3 | 9.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.3 | 19.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 1.3 | 16.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 1.2 | 66.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 1.2 | 20.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 1.2 | 15.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.2 | 37.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 1.1 | 56.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 1.1 | 28.0 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 1.1 | 60.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 1.1 | 35.3 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 1.0 | 75.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 1.0 | 26.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 1.0 | 29.9 | REACTOME SIGNAL AMPLIFICATION | Genes involved in Signal amplification |
| 1.0 | 26.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.9 | 18.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.9 | 20.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.9 | 15.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.8 | 9.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.8 | 9.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.7 | 10.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.6 | 10.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.6 | 5.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 4.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.6 | 21.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.6 | 18.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.5 | 8.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.5 | 16.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.5 | 11.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.5 | 3.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.4 | 5.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.4 | 6.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.4 | 12.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.4 | 7.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.4 | 6.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.4 | 10.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 20.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 3.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 16.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 5.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 3.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 4.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 7.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 9.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 5.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.3 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.1 | 1.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.9 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |