Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Dmc1
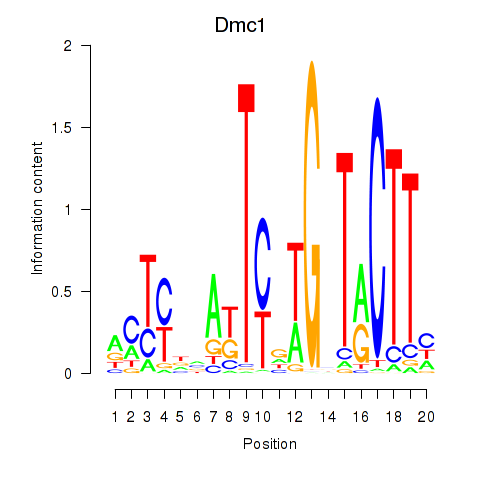
Z-value: 1.60
Transcription factors associated with Dmc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dmc1
|
ENSRNOG00000013807 | DNA meiotic recombinase 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dmc1 | rn6_v1_chr7_-_120839577_120839577 | -0.01 | 9.0e-01 | Click! |
Activity profile of Dmc1 motif
Sorted Z-values of Dmc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_76722965 | 83.54 |
ENSRNOT00000052129
|
LOC100912485
|
alcohol sulfotransferase-like |
| chr14_+_22375955 | 71.87 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr1_+_65576535 | 64.34 |
ENSRNOT00000026575
|
Slc27a5
|
solute carrier family 27 member 5 |
| chr19_+_487723 | 60.99 |
ENSRNOT00000061734
|
Ces2j
|
carboxylesterase 2J |
| chr1_-_258877045 | 59.75 |
ENSRNOT00000071633
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr4_+_154391647 | 57.97 |
ENSRNOT00000081488
ENSRNOT00000079192 |
Mug1
|
murinoglobulin 1 |
| chr17_+_8489266 | 53.61 |
ENSRNOT00000016252
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr14_+_22937421 | 53.20 |
ENSRNOT00000065079
|
RGD1559459
|
similar to Expressed sequence AI788959 |
| chr1_-_258766881 | 53.07 |
ENSRNOT00000015801
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12 |
| chr1_-_148119857 | 52.00 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr14_+_22806132 | 51.46 |
ENSRNOT00000002728
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr4_+_99063181 | 51.16 |
ENSRNOT00000008840
|
Fabp1
|
fatty acid binding protein 1 |
| chr1_+_248723397 | 47.34 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr16_+_18690246 | 46.06 |
ENSRNOT00000081484
|
Mat1a
|
methionine adenosyltransferase 1A |
| chr7_-_15190796 | 46.01 |
ENSRNOT00000006533
|
Cyp4f1
|
cytochrome P450, family 4, subfamily f, polypeptide 1 |
| chr14_+_22517774 | 44.15 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr1_+_107344904 | 43.06 |
ENSRNOT00000082582
|
Gas2
|
growth arrest-specific 2 |
| chr8_-_111761871 | 43.03 |
ENSRNOT00000056483
|
RGD1310507
|
similar to RIKEN cDNA 1300017J02 |
| chr7_+_28066635 | 42.11 |
ENSRNOT00000005844
|
Pah
|
phenylalanine hydroxylase |
| chr16_+_6970342 | 42.06 |
ENSRNOT00000061294
ENSRNOT00000048344 |
Itih4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr20_-_14193690 | 41.11 |
ENSRNOT00000058237
|
Upb1
|
beta-ureidopropionase 1 |
| chr16_+_54164431 | 40.94 |
ENSRNOT00000090763
|
Fgl1
|
fibrinogen-like 1 |
| chr1_+_83103925 | 40.65 |
ENSRNOT00000047540
ENSRNOT00000028196 |
Cyp2b2
|
cytochrome P450, family 2, subfamily b, polypeptide 2 |
| chr1_+_189359853 | 40.02 |
ENSRNOT00000055083
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr5_-_19368431 | 39.75 |
ENSRNOT00000012819
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr1_-_266428239 | 38.75 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr9_+_16924520 | 37.84 |
ENSRNOT00000025094
|
Slc22a7
|
solute carrier family 22 member 7 |
| chr6_+_127946686 | 37.82 |
ENSRNOT00000082680
|
LOC500712
|
Ab1-233 |
| chr1_-_259287684 | 37.00 |
ENSRNOT00000054724
|
Cyp2c22
|
cytochrome P450, family 2, subfamily c, polypeptide 22 |
| chr2_-_23256158 | 36.58 |
ENSRNOT00000015336
|
Bhmt
|
betaine-homocysteine S-methyltransferase |
| chr1_+_189289957 | 35.30 |
ENSRNOT00000020587
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr1_+_83163079 | 35.28 |
ENSRNOT00000077725
ENSRNOT00000034845 |
Cyp2b3
|
cytochrome P450, family 2, subfamily b, polypeptide 3 |
| chr4_+_154215250 | 34.01 |
ENSRNOT00000072465
|
Mug2
|
murinoglobulin 2 |
| chr17_+_78793336 | 33.23 |
ENSRNOT00000057898
|
Mt1
|
metallothionein 1 |
| chr16_+_18690649 | 32.51 |
ENSRNOT00000015190
|
Mat1a
|
methionine adenosyltransferase 1A |
| chr2_+_20857202 | 32.13 |
ENSRNOT00000078919
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr4_-_115332052 | 30.53 |
ENSRNOT00000017643
|
Clec4f
|
C-type lectin domain family 4, member F |
| chr6_-_104631355 | 30.36 |
ENSRNOT00000007825
|
Slc10a1
|
solute carrier family 10 member 1 |
| chr2_-_243407608 | 29.88 |
ENSRNOT00000014631
|
Mttp
|
microsomal triglyceride transfer protein |
| chr2_+_235264219 | 28.67 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr12_-_22194287 | 28.52 |
ENSRNOT00000082895
ENSRNOT00000001905 |
Tfr2
|
transferrin receptor 2 |
| chr1_+_83933942 | 28.15 |
ENSRNOT00000068690
|
Cyp2f4
|
cytochrome P450, family 2, subfamily f, polypeptide 4 |
| chr17_-_14949064 | 28.11 |
ENSRNOT00000079489
ENSRNOT00000081454 |
AABR07027128.1
|
|
| chr16_-_81797815 | 27.78 |
ENSRNOT00000026666
|
Proz
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr1_-_263803150 | 26.56 |
ENSRNOT00000017840
|
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr9_-_4879755 | 26.15 |
ENSRNOT00000047615
|
RGD1559960
|
similar to Sulfotransferase K1 (rSULT1C2) |
| chr10_-_98544447 | 26.03 |
ENSRNOT00000073149
|
Abca6
|
ATP binding cassette subfamily A member 6 |
| chr14_+_22251499 | 25.58 |
ENSRNOT00000087991
ENSRNOT00000002705 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr5_-_32956159 | 25.56 |
ENSRNOT00000078264
|
Cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr3_-_148073554 | 25.49 |
ENSRNOT00000055407
|
Defb27
|
defensin beta 27 |
| chr1_+_83653234 | 25.10 |
ENSRNOT00000085008
ENSRNOT00000084230 ENSRNOT00000090071 |
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr17_-_69460321 | 24.68 |
ENSRNOT00000058367
|
Akr1c1
|
aldo-keto reductase family 1, member C1 |
| chr2_+_182006242 | 24.54 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr7_+_18310624 | 23.73 |
ENSRNOT00000075258
|
Actl9
|
actin-like 9 |
| chr2_+_54466280 | 23.72 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chrX_-_143558521 | 23.41 |
ENSRNOT00000056598
|
LOC688842
|
hypothetical protein LOC688842 |
| chr12_+_19196611 | 22.71 |
ENSRNOT00000001801
|
Azgp1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr8_+_52729003 | 22.67 |
ENSRNOT00000081738
|
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr5_+_151776004 | 22.63 |
ENSRNOT00000009683
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr2_+_150146234 | 22.03 |
ENSRNOT00000018761
|
Aadac
|
arylacetamide deacetylase |
| chr9_+_100281339 | 21.97 |
ENSRNOT00000029127
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr5_+_124476168 | 21.79 |
ENSRNOT00000077754
|
RGD1564074
|
similar to novel protein |
| chr1_-_241460868 | 21.47 |
ENSRNOT00000054776
|
RGD1560242
|
similar to RIKEN cDNA 1700028P14 |
| chrX_-_65400298 | 21.02 |
ENSRNOT00000032121
|
Vsig4
|
V-set and immunoglobulin domain containing 4 |
| chr1_-_89539210 | 20.97 |
ENSRNOT00000077462
ENSRNOT00000028644 |
Hpn
|
hepsin |
| chr12_+_24158766 | 20.80 |
ENSRNOT00000001963
|
Ccl26
|
C-C motif chemokine ligand 26 |
| chr2_+_114423533 | 20.76 |
ENSRNOT00000091221
|
Slc2a2
|
solute carrier family 2 member 2 |
| chr14_+_22142364 | 20.64 |
ENSRNOT00000002699
|
Sult1b1
|
sulfotransferase family 1B member 1 |
| chrX_-_79066932 | 20.60 |
ENSRNOT00000057460
|
RGD1561552
|
similar to WASP family 1 |
| chr16_+_50181316 | 20.49 |
ENSRNOT00000077662
|
F11
|
coagulation factor XI |
| chrX_-_23139694 | 20.37 |
ENSRNOT00000033656
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr9_+_95274707 | 20.28 |
ENSRNOT00000045163
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr7_+_51794173 | 20.26 |
ENSRNOT00000043774
|
Otogl
|
otogelin-like |
| chr17_-_43675934 | 20.16 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr9_-_113246545 | 20.06 |
ENSRNOT00000034759
|
Tmem232
|
transmembrane protein 232 |
| chr1_-_164142206 | 19.97 |
ENSRNOT00000081669
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr7_-_20118466 | 19.84 |
ENSRNOT00000080523
|
RGD1565071
|
similar to hypothetical protein 4930509O22 |
| chrX_-_153457040 | 19.81 |
ENSRNOT00000084905
|
LOC689600
|
hypothetical protein LOC689600 |
| chr7_+_143629455 | 19.77 |
ENSRNOT00000073951
|
Krt18
|
keratin 18 |
| chr1_+_229519506 | 19.73 |
ENSRNOT00000016593
|
Glyatl2
|
glycine-N-acyltransferase-like 2 |
| chrX_-_14783792 | 19.64 |
ENSRNOT00000087609
|
LOC691895
|
similar to ferritin, heavy polypeptide-like 17 |
| chr1_-_162385575 | 19.46 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chrX_+_22674759 | 19.44 |
ENSRNOT00000004683
|
LOC685789
|
hypothetical protein LOC685789 |
| chr3_+_143129248 | 19.23 |
ENSRNOT00000006667
|
Cst8
|
cystatin 8 |
| chr16_-_49003246 | 18.98 |
ENSRNOT00000089501
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr11_-_69355854 | 18.92 |
ENSRNOT00000002975
|
Ropn1
|
rhophilin associated tail protein 1 |
| chr9_+_95256627 | 18.84 |
ENSRNOT00000025291
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr15_+_36609296 | 18.78 |
ENSRNOT00000010795
|
Rnf17
|
ring finger protein 17 |
| chr16_-_54628458 | 18.59 |
ENSRNOT00000042587
|
Adam24
|
ADAM metallopeptidase domain 24 |
| chr2_+_104290726 | 18.36 |
ENSRNOT00000017387
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr1_-_84834039 | 18.30 |
ENSRNOT00000088960
|
AABR07002782.1
|
|
| chr1_-_167588630 | 18.25 |
ENSRNOT00000050707
|
Olr39
|
olfactory receptor 39 |
| chr1_-_89543967 | 18.12 |
ENSRNOT00000079631
|
Hpn
|
hepsin |
| chr5_-_39611053 | 17.95 |
ENSRNOT00000046595
|
Fhl5
|
four and a half LIM domains 5 |
| chrX_+_110233170 | 17.89 |
ENSRNOT00000036019
|
4930513O06Rik
|
RIKEN cDNA 4930513O06 gene |
| chr6_-_10891146 | 17.85 |
ENSRNOT00000020328
|
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr2_-_188413219 | 17.78 |
ENSRNOT00000065065
|
Fdps
|
farnesyl diphosphate synthase |
| chr9_+_95295701 | 17.74 |
ENSRNOT00000025045
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr1_+_102900286 | 17.66 |
ENSRNOT00000017468
|
Ldha
|
lactate dehydrogenase A |
| chr17_+_76008807 | 17.60 |
ENSRNOT00000070895
|
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr1_-_81534140 | 17.53 |
ENSRNOT00000027202
|
Tex101
|
testis expressed 101 |
| chr17_+_81352700 | 17.51 |
ENSRNOT00000024736
|
Mrc1
|
mannose receptor, C type 1 |
| chr15_-_45550285 | 17.38 |
ENSRNOT00000012948
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr3_-_7141522 | 17.37 |
ENSRNOT00000014572
|
Cel
|
carboxyl ester lipase |
| chr1_-_198486157 | 17.33 |
ENSRNOT00000022758
|
Zg16
|
zymogen granule protein 16 |
| chr9_-_32868371 | 17.29 |
ENSRNOT00000038369
|
LOC689725
|
similar to chromosome 9 open reading frame 79 |
| chr10_+_109950988 | 17.16 |
ENSRNOT00000071293
|
LOC688321
|
similar to Lung carbonyl reductase [NADPH] (NADPH-dependent carbonyl reductase) (LCR) (Adipocyte P27 protein) (AP27) |
| chr14_+_84274550 | 17.15 |
ENSRNOT00000006053
ENSRNOT00000087383 |
Sec14l4
|
SEC14-like lipid binding 4 |
| chr5_-_75116490 | 17.10 |
ENSRNOT00000042788
|
Txndc8
|
thioredoxin domain containing 8 |
| chr2_-_210749991 | 16.98 |
ENSRNOT00000051261
ENSRNOT00000052403 |
Gstm6l
|
glutathione S-transferase, mu 6-like |
| chr15_-_57651041 | 16.79 |
ENSRNOT00000072138
|
Spert
|
spermatid associated |
| chr15_+_80040842 | 16.79 |
ENSRNOT00000043065
|
RGD1306441
|
similar to RIKEN cDNA 4921530L21 |
| chr16_-_24951612 | 16.65 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr4_-_62840357 | 16.61 |
ENSRNOT00000059892
|
Slc13a4
|
solute carrier family 13 member 4 |
| chr10_+_92245442 | 16.53 |
ENSRNOT00000006808
|
Sppl2c
|
signal peptide peptidase like 2C |
| chr2_-_132301073 | 16.46 |
ENSRNOT00000058288
|
RGD1563562
|
similar to GTPase activating protein testicular GAP1 |
| chr20_-_10386663 | 16.42 |
ENSRNOT00000045275
ENSRNOT00000042432 |
Cbs
|
cystathionine beta synthase |
| chr4_-_176026133 | 16.20 |
ENSRNOT00000043374
ENSRNOT00000046598 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr6_+_104718512 | 15.93 |
ENSRNOT00000007947
|
Smoc1
|
SPARC related modular calcium binding 1 |
| chr16_-_39970532 | 15.79 |
ENSRNOT00000071331
|
Spata4
|
spermatogenesis associated 4 |
| chr6_+_111180108 | 15.66 |
ENSRNOT00000082027
|
Gstz1
|
glutathione S-transferase zeta 1 |
| chr4_+_70776046 | 15.64 |
ENSRNOT00000040403
|
Prss1
|
protease, serine 1 |
| chr17_+_15194262 | 15.63 |
ENSRNOT00000073070
|
AABR07072539.1
|
|
| chrX_+_83570346 | 15.63 |
ENSRNOT00000045603
|
Ube2d4l1
|
ubiquitin-conjugating enzyme E2D 4 like 1 |
| chr1_-_198267093 | 15.57 |
ENSRNOT00000047477
|
RGD1563217
|
similar to RIKEN cDNA 4930451I11 |
| chr19_+_52077501 | 15.40 |
ENSRNOT00000079240
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr18_-_45380730 | 15.24 |
ENSRNOT00000037904
|
Pudp
|
pseudouridine 5'-phosphatase |
| chr1_+_167937026 | 15.22 |
ENSRNOT00000020655
|
Olr57
|
olfactory receptor 57 |
| chr10_+_83954279 | 15.11 |
ENSRNOT00000006594
|
Ttll6
|
tubulin tyrosine ligase like 6 |
| chr3_-_3266260 | 15.10 |
ENSRNOT00000034538
|
Glt6d1
|
glycosyltransferase 6 domain containing 1 |
| chr1_-_107385257 | 14.97 |
ENSRNOT00000074296
|
Ccdc179
|
coiled-coil domain containing 179 |
| chr9_-_79630452 | 14.88 |
ENSRNOT00000078125
ENSRNOT00000086044 ENSRNOT00000089283 |
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr4_+_52129556 | 14.82 |
ENSRNOT00000009435
|
Hyal6
|
hyaluronoglucosaminidase 6 |
| chr16_-_54533871 | 14.74 |
ENSRNOT00000015144
|
LOC100910977
|
disintegrin and metalloproteinase domain-containing protein 25-like |
| chr4_+_10295122 | 14.66 |
ENSRNOT00000017374
|
Ccdc146
|
coiled-coil domain containing 146 |
| chr4_-_161091010 | 14.61 |
ENSRNOT00000043708
|
Senp17
|
Sumo1/sentrin/SMT3 specific peptidase 17 |
| chr12_-_18274515 | 14.59 |
ENSRNOT00000078075
|
LOC100361826
|
sialophorin-like |
| chrX_+_90588124 | 14.59 |
ENSRNOT00000045685
|
Tgif2lx2
|
TGFB-induced factor homeobox 2-like, X-linked 2 |
| chr4_+_157107469 | 14.58 |
ENSRNOT00000015678
|
C1rl
|
complement C1r subcomponent like |
| chrX_-_139263974 | 14.56 |
ENSRNOT00000050147
|
MGC114492
|
similar to melanoma antigen family A, 5 |
| chr2_-_185303610 | 14.51 |
ENSRNOT00000093479
ENSRNOT00000046286 |
NEWGENE_1592020
|
protease, serine 48 |
| chr17_-_42127678 | 14.42 |
ENSRNOT00000024196
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chrX_-_98095663 | 14.38 |
ENSRNOT00000004491
|
Rbm31y
|
RNA binding motif 31, Y-linked |
| chr9_+_35289443 | 14.36 |
ENSRNOT00000037765
ENSRNOT00000060503 |
Spata31e1
|
SPATA31 subfamily E, member 1 |
| chr1_-_102780381 | 14.23 |
ENSRNOT00000080132
|
Saa4
|
serum amyloid A4 |
| chr7_-_54855557 | 14.17 |
ENSRNOT00000039475
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1 |
| chr16_+_51730452 | 13.96 |
ENSRNOT00000078614
|
Adam34l
|
a disintegrin and metalloprotease domain 34-like |
| chr7_+_140781799 | 13.93 |
ENSRNOT00000087932
|
Dnajc22
|
DnaJ heat shock protein family (Hsp40) member C22 |
| chr1_+_196737627 | 13.90 |
ENSRNOT00000066652
|
LOC100271845
|
hypothetical protein LOC100271845 |
| chr1_-_227359809 | 13.90 |
ENSRNOT00000088080
|
1700025F22Rik
|
RIKEN cDNA 1700025F22 gene |
| chr1_-_153096269 | 13.78 |
ENSRNOT00000022578
|
Tmem135
|
transmembrane protein 135 |
| chrX_-_152367861 | 13.68 |
ENSRNOT00000089414
|
LOC103690371
|
melanoma-associated antigen 10-like |
| chr8_+_4085829 | 13.67 |
ENSRNOT00000031286
|
Actl9b
|
actin-like 9b |
| chr15_-_5467047 | 13.65 |
ENSRNOT00000042743
|
Spetex-2F
|
Spetex-2F protein |
| chrX_-_135761025 | 13.59 |
ENSRNOT00000033044
|
RGD1564955
|
similar to fibrous sheath interacting protein 2 |
| chr8_+_81766041 | 13.59 |
ENSRNOT00000032280
|
Onecut1
|
one cut homeobox 1 |
| chr6_+_127282466 | 13.49 |
ENSRNOT00000012124
|
Otub2
|
OTU deubiquitinase, ubiquitin aldehyde binding 2 |
| chr11_+_88095170 | 13.29 |
ENSRNOT00000041557
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr1_+_91363492 | 13.13 |
ENSRNOT00000014517
|
Cebpa
|
CCAAT/enhancer binding protein alpha |
| chr6_-_107461041 | 13.11 |
ENSRNOT00000058127
|
Heatr4
|
HEAT repeat containing 4 |
| chr14_+_85814824 | 13.04 |
ENSRNOT00000090046
ENSRNOT00000083756 |
Ankrd36
|
ankyrin repeat domain 36 |
| chr14_-_108284619 | 13.02 |
ENSRNOT00000078814
|
LOC690352
|
hypothetical protein LOC690352 |
| chrX_+_97074710 | 12.92 |
ENSRNOT00000044379
|
RGD1561230
|
similar to RIKEN cDNA 4921511C20 gene |
| chr1_-_81550598 | 12.90 |
ENSRNOT00000051671
|
RGD1564380
|
similar to BC049730 protein |
| chr4_-_160803558 | 12.85 |
ENSRNOT00000049542
|
Senp18
|
Sumo1/sentrin/SMT3 specific peptidase 18 |
| chr5_+_162127810 | 12.79 |
ENSRNOT00000038858
|
Pramef27
|
PRAME family member 27 |
| chr9_+_4107246 | 12.73 |
ENSRNOT00000078212
|
AABR07066160.1
|
|
| chr5_+_165415136 | 12.70 |
ENSRNOT00000016317
ENSRNOT00000079407 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr9_+_95285592 | 12.70 |
ENSRNOT00000063853
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chrX_+_130007087 | 12.58 |
ENSRNOT00000074996
|
Tex13c
|
TEX13 family member C |
| chr9_+_95233957 | 12.55 |
ENSRNOT00000071003
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr1_+_101603222 | 12.46 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr17_+_89452814 | 12.26 |
ENSRNOT00000058760
|
RGD1561231
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase like 1) |
| chr4_-_6449949 | 12.19 |
ENSRNOT00000062026
|
Galntl5
|
polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr1_+_282694906 | 12.19 |
ENSRNOT00000074303
|
Ces2c
|
carboxylesterase 2C |
| chr10_+_88620655 | 12.18 |
ENSRNOT00000055248
|
Hspb9
|
heat shock protein family B (small) member 9 |
| chr20_-_11764233 | 12.08 |
ENSRNOT00000001638
|
Pttg1ip
|
pituitary tumor-transforming 1 interacting protein |
| chr14_-_108285008 | 12.06 |
ENSRNOT00000087501
|
LOC690352
|
hypothetical protein LOC690352 |
| chr1_-_104024682 | 11.94 |
ENSRNOT00000056081
|
Mrgprx1
|
MAS-related GPR, member X1 |
| chr4_-_30556814 | 11.91 |
ENSRNOT00000012760
|
Pdk4
|
pyruvate dehydrogenase kinase 4 |
| chr6_+_2886764 | 11.86 |
ENSRNOT00000023114
|
Ttc39d
|
tetratricopeptide repeat domain 39D |
| chr2_-_137153551 | 11.86 |
ENSRNOT00000083443
|
AABR07010476.1
|
|
| chr3_-_14571640 | 11.68 |
ENSRNOT00000060013
|
Ggta1l1
|
glycoprotein, alpha-galactosyltransferase 1-like 1 |
| chr10_-_75202030 | 11.67 |
ENSRNOT00000050349
|
Olr1522
|
olfactory receptor 1522 |
| chr1_+_102924059 | 11.67 |
ENSRNOT00000078137
|
AC099150.1
|
|
| chr16_+_72011870 | 11.65 |
ENSRNOT00000092198
|
Adam5
|
ADAM metallopeptidase domain 5 |
| chr10_+_34414854 | 11.65 |
ENSRNOT00000074980
|
LOC684471
|
similar to olfactory receptor 1394 |
| chrX_-_61151601 | 11.59 |
ENSRNOT00000004132
|
Mageb4
|
melanoma antigen family B, 4 |
| chr15_+_51034620 | 11.46 |
ENSRNOT00000032826
|
Nkx2-6
|
NK2 homeobox 6 |
| chr2_-_196415530 | 11.40 |
ENSRNOT00000064238
|
RGD1359334
|
similar to hypothetical protein FLJ20519 |
| chr10_+_59894340 | 11.38 |
ENSRNOT00000080446
|
Spata22
|
spermatogenesis associated 22 |
| chr1_+_190462327 | 11.37 |
ENSRNOT00000030732
|
LOC691519
|
similar to ankyrin repeat domain 26 |
| chr6_+_84293402 | 11.35 |
ENSRNOT00000072657
|
LOC108351365
|
uncharacterized LOC108351365 |
| chr6_-_108167185 | 11.35 |
ENSRNOT00000015545
|
Aldh6a1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr10_-_105412627 | 11.35 |
ENSRNOT00000034817
|
Qrich2
|
glutamine rich 2 |
| chr10_+_87782376 | 11.32 |
ENSRNOT00000017415
|
LOC680396
|
hypothetical protein LOC680396 |
| chr16_+_51748970 | 11.29 |
ENSRNOT00000059182
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A |
| chr7_-_22943159 | 11.22 |
ENSRNOT00000047547
|
Vom2r57
|
vomeronasal 2 receptor, 57 |
| chr15_+_18322327 | 11.13 |
ENSRNOT00000009650
|
Fam3d
|
family with sequence similarity 3, member D |
| chr8_-_115274165 | 11.12 |
ENSRNOT00000056386
|
LOC102550160
|
IQ domain-containing protein F5-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Dmc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.1 | 51.2 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 16.1 | 64.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 15.7 | 78.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 13.7 | 369.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 13.0 | 39.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 12.9 | 38.7 | GO:0010034 | response to acetate(GO:0010034) |
| 12.2 | 36.6 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 12.1 | 48.5 | GO:0019483 | beta-alanine biosynthetic process(GO:0019483) |
| 9.8 | 29.3 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 9.6 | 38.4 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 9.3 | 120.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 7.9 | 23.7 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 7.8 | 23.4 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 7.2 | 57.8 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 6.9 | 27.6 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 5.7 | 11.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 5.5 | 33.2 | GO:0046687 | response to chromate(GO:0046687) |
| 4.9 | 24.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 4.8 | 14.4 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) cellular response to iron(II) ion(GO:0071282) |
| 4.6 | 18.6 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 4.6 | 63.8 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 4.3 | 60.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 4.3 | 29.9 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 4.2 | 20.8 | GO:0009758 | carbohydrate utilization(GO:0009758) fructose transport(GO:0015755) |
| 4.0 | 20.0 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 4.0 | 55.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 3.6 | 7.2 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 3.3 | 6.7 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 3.3 | 16.6 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 3.2 | 6.4 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 3.1 | 9.4 | GO:0061228 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 3.0 | 9.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 3.0 | 11.9 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 2.9 | 17.7 | GO:0019659 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 2.9 | 20.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 2.9 | 20.4 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 2.9 | 8.7 | GO:2000424 | T-helper 1 cell lineage commitment(GO:0002296) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 2.9 | 22.8 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 2.8 | 2.8 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 2.8 | 30.5 | GO:0051132 | NK T cell activation(GO:0051132) |
| 2.8 | 22.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 2.7 | 10.9 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 2.7 | 2.7 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 2.7 | 19.0 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 2.7 | 8.0 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 2.6 | 10.6 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 2.6 | 7.9 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 2.6 | 7.9 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 2.6 | 36.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 2.6 | 17.9 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 2.5 | 7.6 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 2.4 | 26.8 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 2.4 | 7.1 | GO:1902959 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 2.3 | 9.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 2.2 | 6.6 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 2.2 | 6.6 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 2.2 | 8.7 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 2.2 | 28.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 2.1 | 42.7 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 2.1 | 18.8 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 2.0 | 6.1 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 2.0 | 6.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 1.9 | 5.8 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 1.9 | 11.6 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 1.8 | 11.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 1.8 | 98.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 1.7 | 1.7 | GO:1904444 | regulation of establishment of Sertoli cell barrier(GO:1904444) |
| 1.7 | 9.9 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 1.5 | 13.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.5 | 4.6 | GO:0043132 | phospholipid transfer to membrane(GO:0006649) NAD transport(GO:0043132) regulation of bleb assembly(GO:1904170) |
| 1.5 | 16.8 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 1.5 | 7.5 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 1.5 | 16.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 1.4 | 36.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 1.4 | 121.9 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 1.4 | 22.8 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 1.4 | 14.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.4 | 4.2 | GO:0046099 | guanine salvage(GO:0006178) GMP catabolic process(GO:0046038) guanine biosynthetic process(GO:0046099) |
| 1.4 | 56.1 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 1.4 | 4.2 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 1.4 | 11.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 1.4 | 6.9 | GO:0009624 | response to nematode(GO:0009624) |
| 1.4 | 51.1 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 1.4 | 15.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 1.4 | 5.4 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 1.3 | 28.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.3 | 5.3 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 1.3 | 5.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 1.3 | 42.0 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 1.3 | 5.2 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 1.3 | 7.7 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 1.3 | 34.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 1.2 | 22.1 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 1.2 | 3.5 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 1.1 | 2.3 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 1.1 | 17.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 1.1 | 8.5 | GO:0032571 | response to vitamin K(GO:0032571) |
| 1.1 | 6.4 | GO:0051182 | coenzyme transport(GO:0051182) |
| 1.0 | 3.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 1.0 | 11.4 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 1.0 | 8.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 1.0 | 4.8 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.9 | 8.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.9 | 3.7 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.9 | 2.7 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.9 | 3.6 | GO:0033026 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.9 | 5.5 | GO:2000847 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.9 | 1.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.9 | 7.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.9 | 0.9 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.9 | 2.6 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.8 | 5.8 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.8 | 5.0 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.8 | 5.8 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.8 | 3.2 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.8 | 10.3 | GO:0071468 | cellular response to acidic pH(GO:0071468) |
| 0.8 | 18.4 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.8 | 6.9 | GO:0033504 | floor plate development(GO:0033504) |
| 0.7 | 8.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.7 | 2.2 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.7 | 5.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.7 | 7.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.7 | 10.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.7 | 3.6 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.7 | 12.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.7 | 2.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.7 | 8.4 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.7 | 5.5 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.7 | 6.9 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.7 | 4.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.6 | 4.5 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.6 | 9.0 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.6 | 2.6 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.6 | 2.5 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.6 | 3.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.6 | 2.5 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.6 | 2.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.6 | 3.6 | GO:1904117 | cellular response to vasopressin(GO:1904117) |
| 0.6 | 2.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.6 | 6.5 | GO:0010603 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.6 | 3.0 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) gamma-aminobutyric acid receptor clustering(GO:0097112) receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.6 | 2.9 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.6 | 7.9 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.6 | 1.7 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.6 | 5.5 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.6 | 7.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.5 | 2.7 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.5 | 12.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.5 | 2.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.5 | 2.7 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.5 | 26.8 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.5 | 4.7 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.5 | 10.4 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.5 | 5.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.5 | 5.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.5 | 1.5 | GO:1900114 | positive regulation of histone H3-K9 dimethylation(GO:1900111) positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.5 | 17.5 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.5 | 4.3 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.5 | 3.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.5 | 4.6 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.5 | 4.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.5 | 5.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.4 | 4.9 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.4 | 3.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.4 | 3.0 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.4 | 0.8 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.4 | 2.5 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.4 | 3.3 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.4 | 2.5 | GO:1990928 | cellular response to rapamycin(GO:0072752) response to amino acid starvation(GO:1990928) |
| 0.4 | 1.2 | GO:0044726 | protection of DNA demethylation of female pronucleus(GO:0044726) |
| 0.4 | 2.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.4 | 4.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 | 15.6 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.4 | 3.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.4 | 4.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 2.3 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.4 | 1.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.4 | 3.0 | GO:0030823 | regulation of cGMP metabolic process(GO:0030823) |
| 0.4 | 1.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 7.8 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.4 | 4.5 | GO:0046068 | cGMP metabolic process(GO:0046068) |
| 0.4 | 4.8 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.4 | 2.2 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.4 | 7.7 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.4 | 17.1 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.4 | 1.4 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.4 | 2.5 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.4 | 2.5 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.3 | 3.1 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.3 | 2.8 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.3 | 9.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.3 | 2.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.3 | 3.9 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 6.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.3 | 2.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 3.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.3 | 4.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 4.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 2.1 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.3 | 3.8 | GO:0072711 | cellular response to hydroxyurea(GO:0072711) |
| 0.3 | 1.8 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.3 | 2.0 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.3 | 364.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.3 | 11.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.3 | 1.6 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.3 | 4.0 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.3 | 2.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.3 | 2.8 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.3 | 5.6 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.3 | 1.3 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.3 | 0.8 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 0.3 | 4.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 7.7 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.2 | 6.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 3.6 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.2 | 4.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 4.4 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 0.7 | GO:2000523 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 1.8 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.2 | 4.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 6.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.2 | 0.9 | GO:0051692 | cellular oligosaccharide metabolic process(GO:0051691) cellular oligosaccharide catabolic process(GO:0051692) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.2 | 2.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 3.8 | GO:2000778 | positive regulation of interleukin-6 secretion(GO:2000778) |
| 0.2 | 1.5 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.2 | 1.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 13.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 3.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.2 | 12.2 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.2 | 0.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 2.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 6.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.2 | 0.8 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.2 | 0.6 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.2 | 0.6 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 1.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.2 | 1.6 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 1.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 0.9 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.2 | 1.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 3.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 12.0 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 1.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 2.1 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.2 | 1.6 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.2 | 3.6 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.2 | 1.2 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.1 | 7.9 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 4.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.9 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.0 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 1.3 | GO:0090084 | chaperone cofactor-dependent protein refolding(GO:0070389) negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 3.7 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 3.1 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 1.1 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 1.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 3.9 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.9 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.1 | 0.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 2.0 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 2.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 4.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 15.2 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 3.0 | GO:0032873 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 0.4 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.1 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 3.4 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 0.9 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 3.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.3 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 2.2 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 1.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 2.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.8 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 1.0 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.1 | 1.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 20.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 1.3 | GO:0019395 | fatty acid oxidation(GO:0019395) |
| 0.1 | 1.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 12.2 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 2.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 7.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 6.4 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 0.9 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 1.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 7.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 0.9 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.5 | GO:1900040 | interleukin-2 secretion(GO:0070970) regulation of interleukin-2 secretion(GO:1900040) |
| 0.1 | 2.4 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) |
| 0.1 | 3.4 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 4.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 2.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.4 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 1.3 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.0 | 0.5 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.6 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.5 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 1.4 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 0.9 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.7 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 1.1 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 1.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.3 | GO:0010453 | regulation of cell fate commitment(GO:0010453) |
| 0.0 | 0.3 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 51.2 | GO:0045179 | apical cortex(GO:0045179) |
| 4.6 | 27.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 4.4 | 13.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 3.4 | 23.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 3.2 | 18.9 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 3.1 | 24.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 2.6 | 18.2 | GO:0060205 | cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 2.5 | 7.6 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 2.5 | 45.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 2.4 | 16.5 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 2.1 | 10.6 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 1.8 | 27.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 1.4 | 4.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 1.4 | 4.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.3 | 6.7 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.3 | 24.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 1.3 | 8.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 1.2 | 3.6 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 1.2 | 28.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 1.2 | 131.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 1.2 | 64.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 1.0 | 4.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 1.0 | 7.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 1.0 | 7.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.9 | 6.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.9 | 7.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.9 | 12.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.9 | 5.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.8 | 3.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.8 | 51.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.8 | 2.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.8 | 55.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.8 | 7.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.7 | 18.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.7 | 15.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.7 | 79.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.7 | 2.2 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.7 | 2.9 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.7 | 2.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.7 | 8.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.7 | 17.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.6 | 1.3 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.6 | 46.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.6 | 6.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.6 | 2.4 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.6 | 3.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.6 | 1.7 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.6 | 4.6 | GO:0032059 | bleb(GO:0032059) |
| 0.6 | 2.3 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.6 | 11.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.6 | 438.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.5 | 2.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.5 | 41.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.5 | 4.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.5 | 4.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.5 | 2.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.5 | 3.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 51.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.4 | 1.8 | GO:0035363 | histone locus body(GO:0035363) |
| 0.4 | 9.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.4 | 2.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.4 | 3.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 7.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 1.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 4.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.3 | 5.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 4.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 0.9 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.3 | 0.9 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 6.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 2.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.3 | 7.6 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 2.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 3.0 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 4.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 14.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 7.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 1.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 1.0 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.2 | 4.7 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.2 | 2.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 11.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 3.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 6.6 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.2 | 7.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 4.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 1.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 0.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 3.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 30.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 8.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 3.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 2.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 2.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 3.2 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.7 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 2.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 4.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 3.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 18.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.0 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 4.9 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 2.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.0 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.1 | 2.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 5.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.0 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 4.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 2.9 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 41.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 19.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 26.2 | 78.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 14.0 | 42.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 12.5 | 413.5 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) |
| 11.1 | 342.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 10.7 | 85.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 9.6 | 115.5 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 9.0 | 36.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 8.2 | 24.7 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 6.4 | 19.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 6.3 | 31.3 | GO:0005534 | galactose binding(GO:0005534) |
| 6.0 | 36.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 6.0 | 18.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 5.8 | 17.4 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 5.0 | 20.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 4.7 | 19.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 4.6 | 55.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 4.6 | 36.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 4.4 | 17.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 4.4 | 22.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 4.3 | 29.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 3.9 | 78.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 3.7 | 14.9 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 3.6 | 10.9 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 3.4 | 13.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 3.3 | 16.6 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 3.3 | 19.7 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 3.2 | 34.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 3.1 | 28.2 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 3.0 | 12.2 | GO:0047619 | acylcarnitine hydrolase activity(GO:0047619) |
| 3.0 | 15.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 3.0 | 45.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 3.0 | 11.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 2.9 | 11.7 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 2.9 | 8.7 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 2.9 | 20.4 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 2.8 | 39.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 2.8 | 19.4 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 2.5 | 10.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 2.5 | 7.4 | GO:0004159 | dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 2.4 | 7.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 2.3 | 20.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 2.2 | 6.7 | GO:0046911 | metal chelating activity(GO:0046911) |
| 2.2 | 6.6 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 2.2 | 8.7 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 2.1 | 6.2 | GO:0071209 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 2.0 | 12.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 2.0 | 16.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 1.8 | 14.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 1.8 | 16.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.8 | 5.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 1.7 | 36.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 1.7 | 13.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.7 | 10.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 1.7 | 5.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.6 | 9.8 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 1.6 | 3.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 1.6 | 6.4 | GO:0042806 | fucose binding(GO:0042806) |
| 1.5 | 6.2 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 1.5 | 4.6 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 1.5 | 4.5 | GO:0047708 | biotinidase activity(GO:0047708) |
| 1.5 | 37.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 1.5 | 14.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 1.5 | 13.1 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 1.4 | 8.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 1.4 | 28.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 1.4 | 8.3 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.3 | 5.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 1.3 | 7.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.3 | 5.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.3 | 6.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 1.3 | 12.7 | GO:0001846 | opsonin binding(GO:0001846) |
| 1.2 | 6.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.2 | 6.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.2 | 3.6 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 1.2 | 7.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.2 | 4.8 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 1.2 | 18.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 1.2 | 12.8 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 1.2 | 3.5 | GO:0004956 | prostaglandin D receptor activity(GO:0004956) |
| 1.2 | 4.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 1.1 | 7.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.1 | 4.4 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 1.1 | 5.4 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 1.0 | 4.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 1.0 | 24.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 1.0 | 6.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 1.0 | 8.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 1.0 | 4.0 | GO:0002046 | opsin binding(GO:0002046) |
| 1.0 | 4.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.0 | 7.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.9 | 107.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.9 | 2.7 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.9 | 4.5 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 0.9 | 3.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.9 | 2.7 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.8 | 11.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.8 | 6.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.8 | 6.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.8 | 14.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.8 | 3.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.8 | 4.7 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.8 | 16.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.8 | 10.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.8 | 6.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.8 | 4.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.8 | 2.3 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 0.8 | 2.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.7 | 20.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.7 | 20.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.7 | 2.9 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.7 | 2.8 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.7 | 6.3 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.7 | 26.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.7 | 17.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.7 | 83.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.7 | 9.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.6 | 133.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.6 | 2.5 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.6 | 2.4 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.6 | 4.7 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.6 | 4.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.6 | 9.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.6 | 8.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.6 | 1.7 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.6 | 4.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.6 | 2.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.6 | 8.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.6 | 1.7 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.5 | 3.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.5 | 2.7 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.5 | 3.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.5 | 36.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.5 | 4.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.5 | 2.5 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.5 | 16.8 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.5 | 3.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.5 | 3.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.5 | 2.8 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.5 | 1.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.4 | 9.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.4 | 4.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.4 | 3.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.4 | 10.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.4 | 2.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.4 | 28.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.4 | 1.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.4 | 1.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.3 | 2.8 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.3 | 7.9 | GO:0048038 | quinone binding(GO:0048038) |
| 0.3 | 1.0 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 8.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 6.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 2.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.3 | 2.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 2.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 1.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 2.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 2.6 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.3 | 0.9 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.3 | 3.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 2.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.3 | 2.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 344.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.3 | 5.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 5.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 2.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.3 | 3.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.3 | 2.6 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.3 | 1.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 5.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 5.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 2.8 | GO:0005536 | glucose binding(GO:0005536) |
| 0.2 | 0.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 6.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 11.7 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.2 | 3.9 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 0.6 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 2.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 10.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.7 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.2 | 1.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 1.0 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 24.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 1.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 3.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.6 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.4 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 10.9 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 1.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 0.8 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 14.3 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 8.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 2.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 5.3 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 3.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 2.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 3.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 4.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.7 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 1.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 2.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 6.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 10.0 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.1 | 2.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 7.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 10.7 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.1 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 9.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 2.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 5.1 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.1 | 1.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 3.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.4 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.1 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.1 | 0.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 2.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 2.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 5.0 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.7 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 1.1 | 18.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.9 | 43.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.7 | 40.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 8.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 2.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 25.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.4 | 35.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.4 | 86.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 7.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.4 | 21.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 18.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.3 | 11.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.3 | 3.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 10.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 5.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 7.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 5.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 4.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 3.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 1.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 4.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 5.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.1 | 172.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 7.9 | 95.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 4.6 | 78.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 4.5 | 131.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 3.7 | 48.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 3.4 | 37.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 2.8 | 27.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 2.3 | 30.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 2.1 | 36.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 2.1 | 25.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 2.1 | 81.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 1.9 | 19.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.8 | 23.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.8 | 9.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 1.7 | 36.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.5 | 43.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.5 | 20.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 1.3 | 29.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 1.0 | 9.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 1.0 | 17.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.9 | 5.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.9 | 35.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.9 | 1.8 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.9 | 20.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.9 | 18.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.7 | 11.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.6 | 16.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 8.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.6 | 6.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.6 | 10.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.6 | 10.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.6 | 8.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.5 | 9.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.5 | 5.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.5 | 9.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.5 | 10.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.5 | 76.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.5 | 5.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.5 | 16.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 5.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 3.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.4 | 2.5 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.4 | 10.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.3 | 3.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 31.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.3 | 4.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 4.6 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.3 | 8.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 4.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 12.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.3 | 18.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 11.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.3 | 5.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 9.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 8.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 11.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 3.2 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.2 | 2.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 3.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.9 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 5.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.9 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.1 | 0.4 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 0.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 2.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |