Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Dlx5_Dlx4
Z-value: 0.70
Transcription factors associated with Dlx5_Dlx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
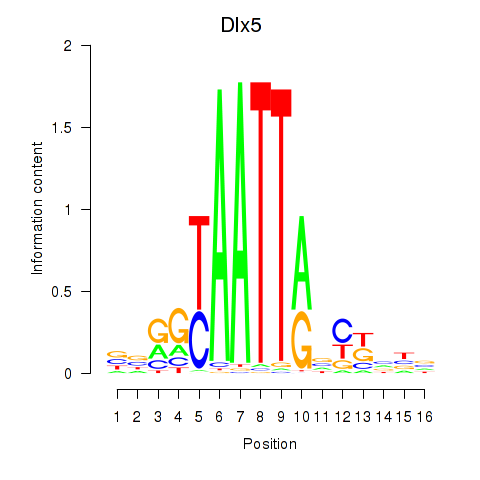
Dlx5
|
ENSRNOG00000010905 | distal-less homeobox 5 |
|
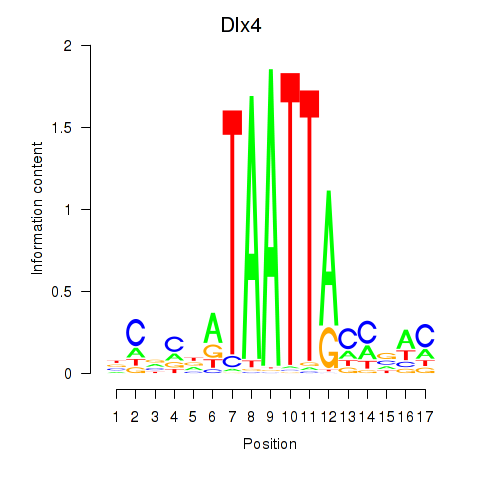
Dlx4
|
ENSRNOG00000004399 | distal-less homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx4 | rn6_v1_chr10_-_82963919_82963919 | -0.35 | 1.5e-10 | Click! |
| Dlx5 | rn6_v1_chr4_-_32392007_32392007 | -0.29 | 8.6e-08 | Click! |
Activity profile of Dlx5_Dlx4 motif
Sorted Z-values of Dlx5_Dlx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_22375955 | 29.58 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr2_-_216382244 | 27.33 |
ENSRNOT00000086695
ENSRNOT00000087259 |
LOC103689940
|
pancreatic alpha-amylase-like |
| chr17_+_72160735 | 26.79 |
ENSRNOT00000038817
|
Itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr2_-_216348194 | 20.13 |
ENSRNOT00000087839
|
LOC103689940
|
pancreatic alpha-amylase-like |
| chr17_+_9736786 | 17.02 |
ENSRNOT00000081920
|
F12
|
coagulation factor XII |
| chr1_+_140998240 | 16.73 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr10_+_96639924 | 16.42 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chr13_-_47397890 | 16.27 |
ENSRNOT00000005505
|
C4bpb
|
complement component 4 binding protein, beta |
| chr6_+_2216623 | 14.48 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr1_-_148119857 | 14.16 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr14_+_22517774 | 13.97 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr1_+_248428099 | 13.70 |
ENSRNOT00000050984
|
Mbl2
|
mannose binding lectin 2 |
| chr2_+_54466280 | 13.30 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr6_+_127946686 | 13.12 |
ENSRNOT00000082680
|
LOC500712
|
Ab1-233 |
| chr17_+_76008807 | 12.97 |
ENSRNOT00000070895
|
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr4_+_154676427 | 12.48 |
ENSRNOT00000019969
ENSRNOT00000087441 |
Mug1
|
murinoglobulin 1 |
| chr5_+_117698764 | 11.96 |
ENSRNOT00000011486
|
Angptl3
|
angiopoietin-like 3 |
| chr1_+_279896973 | 11.74 |
ENSRNOT00000068119
|
Pnliprp2
|
pancreatic lipase related protein 2 |
| chr2_+_200793571 | 11.04 |
ENSRNOT00000091444
|
Hao2
|
hydroxyacid oxidase 2 |
| chr12_+_10636275 | 10.68 |
ENSRNOT00000001285
|
Cyp3a18
|
cytochrome P450, family 3, subfamily a, polypeptide 18 |
| chr14_+_5928737 | 10.06 |
ENSRNOT00000071877
ENSRNOT00000040985 ENSRNOT00000074889 |
Mpa2l
|
macrophage activation 2 like |
| chr3_-_14229067 | 10.02 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr9_-_27447877 | 10.02 |
ENSRNOT00000085195
|
Gsta1
|
glutathione S-transferase alpha 1 |
| chr12_-_19167015 | 9.99 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chrM_+_7758 | 9.96 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr6_-_128003418 | 9.94 |
ENSRNOT00000013896
|
Serpina3c
|
serine (or cysteine) proteinase inhibitor, clade A, member 3C |
| chr9_+_100281339 | 9.31 |
ENSRNOT00000029127
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr13_+_78812394 | 9.30 |
ENSRNOT00000076043
|
Serpinc1
|
serpin family C member 1 |
| chr15_+_57290849 | 9.18 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chr3_+_54253949 | 9.10 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr11_+_74057361 | 9.10 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr20_+_13817795 | 9.05 |
ENSRNOT00000036518
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr1_-_198104109 | 8.85 |
ENSRNOT00000026186
|
Sult1a1
|
sulfotransferase family 1A member 1 |
| chr10_-_98544447 | 8.80 |
ENSRNOT00000073149
|
Abca6
|
ATP binding cassette subfamily A member 6 |
| chr17_-_43537293 | 8.79 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chr2_+_55775274 | 8.71 |
ENSRNOT00000018545
|
C9
|
complement C9 |
| chr14_-_21748356 | 8.64 |
ENSRNOT00000002670
|
Cabs1
|
calcium binding protein, spermatid associated 1 |
| chr2_-_182038178 | 8.25 |
ENSRNOT00000040708
|
Fgb
|
fibrinogen beta chain |
| chr10_+_56662561 | 8.13 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr9_-_80295446 | 7.79 |
ENSRNOT00000023769
|
Tnp1
|
transition protein 1 |
| chr6_-_127816055 | 7.75 |
ENSRNOT00000013175
|
Serpina3m
|
serine (or cysteine) proteinase inhibitor, clade A, member 3M |
| chr10_+_56662242 | 7.72 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr12_+_19196611 | 7.67 |
ENSRNOT00000001801
|
Azgp1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr1_-_258877045 | 7.47 |
ENSRNOT00000071633
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr17_-_69711689 | 7.40 |
ENSRNOT00000041925
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr16_+_74865516 | 7.39 |
ENSRNOT00000058072
|
Atp7b
|
ATPase copper transporting beta |
| chr14_+_96499520 | 7.38 |
ENSRNOT00000074692
|
AABR07016310.1
|
|
| chr9_-_4945352 | 7.32 |
ENSRNOT00000082530
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr3_+_16571602 | 7.28 |
ENSRNOT00000048351
|
LOC100361705
|
rCG64259-like |
| chr4_+_148782479 | 7.12 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr9_+_95161157 | 7.00 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr1_+_213511874 | 6.94 |
ENSRNOT00000078080
ENSRNOT00000016883 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr15_-_42693694 | 6.86 |
ENSRNOT00000022702
|
Gulo
|
gulonolactone (L-) oxidase |
| chr14_+_22142364 | 6.78 |
ENSRNOT00000002699
|
Sult1b1
|
sulfotransferase family 1B member 1 |
| chr11_-_81444375 | 6.74 |
ENSRNOT00000058479
ENSRNOT00000078131 ENSRNOT00000080949 ENSRNOT00000080562 ENSRNOT00000084867 |
Kng1
|
kininogen 1 |
| chr8_+_62779875 | 6.68 |
ENSRNOT00000010831
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr3_+_19366370 | 6.60 |
ENSRNOT00000086557
|
AABR07051689.1
|
|
| chr1_-_164142206 | 6.58 |
ENSRNOT00000081669
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr2_+_223121410 | 6.56 |
ENSRNOT00000087559
|
AABR07013116.1
|
|
| chr20_-_12820466 | 6.42 |
ENSRNOT00000001699
|
Ftcd
|
formimidoyltransferase cyclodeaminase |
| chr11_-_87924816 | 6.37 |
ENSRNOT00000031819
|
Serpind1
|
serpin family D member 1 |
| chr12_+_18679789 | 6.33 |
ENSRNOT00000001863
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr7_+_34326087 | 6.30 |
ENSRNOT00000006971
|
Hal
|
histidine ammonia lyase |
| chr1_+_55219773 | 6.02 |
ENSRNOT00000041610
|
RGD1561667
|
similar to putative protein kinase |
| chr16_-_74864816 | 6.01 |
ENSRNOT00000017164
|
Alg11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr3_+_18970574 | 5.98 |
ENSRNOT00000088394
|
AABR07051665.1
|
|
| chr1_-_258766881 | 5.94 |
ENSRNOT00000015801
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12 |
| chr4_+_98481520 | 5.91 |
ENSRNOT00000078381
ENSRNOT00000048493 |
AABR07060886.1
|
|
| chr10_+_109665682 | 5.90 |
ENSRNOT00000054963
|
Slc25a10
|
solute carrier family 25 member 10 |
| chr14_-_5859581 | 5.80 |
ENSRNOT00000052308
|
AABR07014241.1
|
|
| chr1_+_282567674 | 5.77 |
ENSRNOT00000090543
|
Ces2i
|
carboxylesterase 2I |
| chr16_-_24951612 | 5.66 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr6_+_139405966 | 5.49 |
ENSRNOT00000088974
|
AABR07065693.3
|
|
| chr9_+_61692154 | 5.49 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr15_+_28319136 | 5.44 |
ENSRNOT00000048723
|
Tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr4_-_117575154 | 5.32 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr9_-_114327767 | 5.29 |
ENSRNOT00000085481
|
AABR07068674.1
|
|
| chr3_-_15278645 | 5.23 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr3_+_18315320 | 5.21 |
ENSRNOT00000006954
|
AABR07051626.2
|
|
| chr14_-_79464770 | 5.12 |
ENSRNOT00000008932
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr2_+_140708397 | 5.08 |
ENSRNOT00000088846
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr16_+_50152008 | 5.03 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr1_-_227457629 | 4.95 |
ENSRNOT00000035910
ENSRNOT00000073770 |
Ms4a5
|
membrane spanning 4-domains A5 |
| chr4_-_30338679 | 4.88 |
ENSRNOT00000012050
|
Pon3
|
paraoxonase 3 |
| chr10_-_87286387 | 4.86 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr15_+_28018040 | 4.79 |
ENSRNOT00000041495
|
Rnase4
|
ribonuclease A family member 4 |
| chr3_+_16846412 | 4.79 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chr11_+_88095170 | 4.73 |
ENSRNOT00000041557
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr17_-_69404323 | 4.68 |
ENSRNOT00000051342
ENSRNOT00000066282 |
Akr1c2
|
aldo-keto reductase family 1, member C2 |
| chr4_+_117962319 | 4.64 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr3_+_159936856 | 4.61 |
ENSRNOT00000078703
|
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr9_+_95501778 | 4.56 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr2_-_105089659 | 4.54 |
ENSRNOT00000043381
|
Cpb1
|
carboxypeptidase B1 |
| chr2_+_93758919 | 4.51 |
ENSRNOT00000077782
|
Fabp12
|
fatty acid binding protein 12 |
| chr6_-_41870046 | 4.46 |
ENSRNOT00000005863
|
Lpin1
|
lipin 1 |
| chr8_-_115274165 | 4.45 |
ENSRNOT00000056386
|
LOC102550160
|
IQ domain-containing protein F5-like |
| chr3_+_19045214 | 4.45 |
ENSRNOT00000070878
|
AABR07051670.1
|
|
| chr1_+_229030233 | 4.44 |
ENSRNOT00000084503
|
Glyatl1
|
glycine-N-acyltransferase-like 1 |
| chr17_-_43584152 | 4.42 |
ENSRNOT00000023241
|
Slc17a2
|
solute carrier family 17, member 2 |
| chr1_-_275882444 | 4.38 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr16_+_68586235 | 4.38 |
ENSRNOT00000039592
|
LOC103693984
|
uncharacterized LOC103693984 |
| chr17_-_42127678 | 4.35 |
ENSRNOT00000024196
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr2_+_154921999 | 4.27 |
ENSRNOT00000057620
|
LOC691044
|
similar to GTPase activating protein testicular GAP1 |
| chr2_+_243502073 | 4.26 |
ENSRNOT00000015870
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr8_-_37450340 | 4.26 |
ENSRNOT00000072224
|
LOC100912405
|
urinary protein 3-like |
| chr15_+_18710492 | 4.24 |
ENSRNOT00000012532
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr6_-_128149220 | 4.24 |
ENSRNOT00000014204
|
Gsc
|
goosecoid homeobox |
| chr2_+_26240385 | 4.12 |
ENSRNOT00000024292
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr7_+_23012070 | 4.08 |
ENSRNOT00000042460
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr14_-_2032593 | 4.08 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr8_+_85059051 | 4.00 |
ENSRNOT00000033196
|
Gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr2_+_150146234 | 3.99 |
ENSRNOT00000018761
|
Aadac
|
arylacetamide deacetylase |
| chr7_+_20262680 | 3.97 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr2_+_60131776 | 3.95 |
ENSRNOT00000080786
|
Prlr
|
prolactin receptor |
| chr8_-_8524643 | 3.93 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chr9_-_79630452 | 3.90 |
ENSRNOT00000078125
ENSRNOT00000086044 ENSRNOT00000089283 |
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr7_+_16404755 | 3.89 |
ENSRNOT00000044977
|
LOC108349290
|
olfactory receptor 6C70-like |
| chr9_-_98551410 | 3.82 |
ENSRNOT00000066346
ENSRNOT00000086678 |
Hes6
|
hes family bHLH transcription factor 6 |
| chr19_+_52258947 | 3.76 |
ENSRNOT00000021072
|
Adad2
|
adenosine deaminase domain containing 2 |
| chrX_+_45965301 | 3.75 |
ENSRNOT00000005141
|
Fam47a
|
family with sequence similarity 47, member A |
| chr1_+_22332090 | 3.73 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr3_-_26056818 | 3.72 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr4_-_176528110 | 3.72 |
ENSRNOT00000049569
|
Slco1a2
|
solute carrier organic anion transporter family, member 1A2 |
| chr2_+_200397967 | 3.66 |
ENSRNOT00000025821
|
Reg4
|
regenerating family member 4 |
| chr4_+_1658278 | 3.63 |
ENSRNOT00000073845
|
Olr1250
|
olfactory receptor 1250 |
| chr8_-_56393233 | 3.61 |
ENSRNOT00000016263
|
Fdx1
|
ferredoxin 1 |
| chr6_+_10483308 | 3.57 |
ENSRNOT00000074516
|
Tmem247
|
transmembrane protein 247 |
| chr3_+_18805220 | 3.54 |
ENSRNOT00000071216
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr6_-_138852571 | 3.53 |
ENSRNOT00000081803
|
AABR07065656.8
|
|
| chr19_+_15081590 | 3.53 |
ENSRNOT00000024187
|
Ces1f
|
carboxylesterase 1F |
| chrX_-_23139694 | 3.50 |
ENSRNOT00000033656
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr4_-_155051429 | 3.49 |
ENSRNOT00000020094
|
Klrg1
|
killer cell lectin like receptor G1 |
| chr1_+_264260505 | 3.48 |
ENSRNOT00000018815
|
Wnt8b
|
wingless-type MMTV integration site family, member 8B |
| chr6_-_115616766 | 3.47 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr5_-_12526962 | 3.43 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr3_-_16999720 | 3.42 |
ENSRNOT00000074382
|
RGD1563231
|
similar to immunoglobulin kappa-chain VK-1 |
| chr9_-_105655471 | 3.37 |
ENSRNOT00000014921
|
RGD1560925
|
similar to 2610034M16Rik protein |
| chr8_+_57983556 | 3.37 |
ENSRNOT00000009562
|
RGD1311251
|
similar to RIKEN cDNA 4930550C14 |
| chr16_+_72139063 | 3.33 |
ENSRNOT00000023805
|
Adam18
|
ADAM metallopeptidase domain 18 |
| chr15_-_10455956 | 3.33 |
ENSRNOT00000008289
|
Ngly1
|
N-glycanase 1 |
| chr1_+_167937026 | 3.31 |
ENSRNOT00000020655
|
Olr57
|
olfactory receptor 57 |
| chr6_+_64789940 | 3.31 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr1_-_81596819 | 3.30 |
ENSRNOT00000074350
|
RGD1564380
|
similar to BC049730 protein |
| chr6_-_92643847 | 3.26 |
ENSRNOT00000009183
|
Pygl
|
glycogen phosphorylase L |
| chr4_-_163095614 | 3.25 |
ENSRNOT00000088759
|
RGD1564770
|
similar to CD69 antigen (p60, early T-cell activation antigen) |
| chr1_+_31409579 | 3.22 |
ENSRNOT00000016989
|
RGD1311933
|
similar to RIKEN cDNA 2310057J18 |
| chr6_-_7058314 | 3.21 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr9_-_85243001 | 3.20 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr17_+_69588085 | 3.19 |
ENSRNOT00000064884
|
Akr1c12l1
|
aldo-keto reductase family 1, member C12-like 1 |
| chr6_-_139033819 | 3.19 |
ENSRNOT00000091961
|
AABR07065656.6
|
|
| chr3_-_20419417 | 3.18 |
ENSRNOT00000077772
|
AABR07051733.3
|
|
| chr11_+_85532526 | 3.17 |
ENSRNOT00000036565
|
AABR07034730.2
|
|
| chr8_+_112594691 | 3.16 |
ENSRNOT00000038383
ENSRNOT00000081281 |
Acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr8_-_54994871 | 3.15 |
ENSRNOT00000013014
|
Tex12
|
testis expressed 12 |
| chr13_-_57080491 | 3.14 |
ENSRNOT00000017749
ENSRNOT00000086572 ENSRNOT00000060111 |
Cfh
|
complement factor H |
| chr6_-_139911839 | 3.11 |
ENSRNOT00000077113
ENSRNOT00000084547 |
AABR07065714.1
|
|
| chrM_+_10160 | 3.11 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr5_-_77749613 | 3.08 |
ENSRNOT00000075988
|
Mup5
|
major urinary protein 5 |
| chr3_-_19320915 | 3.08 |
ENSRNOT00000043673
|
RGD1565617
|
similar to Ig variable region, light chain |
| chr5_-_136965191 | 3.08 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr16_-_49820235 | 3.07 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr15_+_28028521 | 3.04 |
ENSRNOT00000089631
|
AC114343.1
|
|
| chr17_+_69634890 | 3.04 |
ENSRNOT00000029049
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr1_+_276240703 | 3.03 |
ENSRNOT00000022126
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr4_+_70689737 | 3.03 |
ENSRNOT00000018852
|
Prss2
|
protease, serine, 2 |
| chrX_+_78769419 | 3.02 |
ENSRNOT00000003190
|
Tbx22
|
T-box 22 |
| chr7_-_129970550 | 3.02 |
ENSRNOT00000055879
|
Mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr13_+_75177965 | 2.98 |
ENSRNOT00000007321
|
Sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr4_+_70977556 | 2.97 |
ENSRNOT00000031984
|
LOC680112
|
hypothetical protein LOC680112 |
| chr1_+_219833299 | 2.97 |
ENSRNOT00000087432
|
Pc
|
pyruvate carboxylase |
| chr6_+_78567970 | 2.96 |
ENSRNOT00000032743
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr1_+_64506735 | 2.96 |
ENSRNOT00000086331
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
| chr7_+_44009069 | 2.95 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr20_-_30947484 | 2.94 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr6_-_140880070 | 2.93 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr3_-_16441030 | 2.92 |
ENSRNOT00000047784
|
AABR07051532.1
|
|
| chr1_+_102924059 | 2.92 |
ENSRNOT00000078137
|
AC099150.1
|
|
| chr1_+_201913148 | 2.90 |
ENSRNOT00000036128
|
Fam24a
|
family with sequence similarity 24, member A |
| chr6_-_143590448 | 2.87 |
ENSRNOT00000056771
|
Ighv8-4
|
immunoglobulin heavy variable V8-4 |
| chr3_-_66417741 | 2.85 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr15_+_33885106 | 2.83 |
ENSRNOT00000024483
|
Dhrs2
|
dehydrogenase/reductase 2 |
| chr4_-_151428894 | 2.82 |
ENSRNOT00000010556
|
Adipor2
|
adiponectin receptor 2 |
| chr20_+_31339787 | 2.82 |
ENSRNOT00000082463
|
Aifm2
|
apoptosis inducing factor, mitochondria associated 2 |
| chr6_-_142372031 | 2.81 |
ENSRNOT00000063929
|
AABR07065814.3
|
|
| chr17_+_15194262 | 2.81 |
ENSRNOT00000073070
|
AABR07072539.1
|
|
| chr6_-_143065639 | 2.80 |
ENSRNOT00000070923
|
AABR07065827.1
|
|
| chr1_+_222861777 | 2.79 |
ENSRNOT00000090872
|
Pla2g16
|
phospholipase A2, group XVI |
| chr20_+_1736377 | 2.79 |
ENSRNOT00000047035
|
Olr1734
|
olfactory receptor 1734 |
| chr19_+_3325893 | 2.77 |
ENSRNOT00000048879
|
RGD1565617
|
similar to Ig variable region, light chain |
| chr10_+_1834518 | 2.76 |
ENSRNOT00000061709
|
Gm1758
|
predicted gene 1758 |
| chr3_-_64543100 | 2.69 |
ENSRNOT00000025803
|
Zfp385b
|
zinc finger protein 385B |
| chr2_-_132551235 | 2.68 |
ENSRNOT00000058250
|
LOC100365525
|
rCG65904-like |
| chr8_-_78233430 | 2.68 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr6_-_141866756 | 2.65 |
ENSRNOT00000068561
|
AABR07065804.1
|
|
| chr4_+_102262007 | 2.65 |
ENSRNOT00000079328
|
AABR07060992.2
|
|
| chr5_-_24926904 | 2.64 |
ENSRNOT00000075288
|
AABR07047232.1
|
|
| chr2_-_24923128 | 2.63 |
ENSRNOT00000044087
|
Pde8b
|
phosphodiesterase 8B |
| chr20_+_1749716 | 2.61 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr10_-_87578854 | 2.60 |
ENSRNOT00000065619
|
LOC680160
|
similar to keratin associated protein 4-7 |
| chr10_-_98124743 | 2.59 |
ENSRNOT00000037415
|
RGD1559578
|
RGD1559578 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Dlx5_Dlx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 28.9 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 6.4 | 25.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 5.7 | 17.0 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 5.4 | 16.3 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 4.2 | 12.7 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 3.7 | 11.0 | GO:0018924 | mandelate metabolic process(GO:0018924) |
| 3.1 | 9.3 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) oxalic acid secretion(GO:0046724) |
| 3.1 | 9.2 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 2.5 | 7.4 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 2.4 | 12.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 2.2 | 15.6 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 2.2 | 4.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 2.1 | 6.3 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 2.1 | 18.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 2.0 | 5.9 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 1.8 | 12.4 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 1.7 | 6.9 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 1.6 | 8.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.6 | 4.7 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.4 | 6.9 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 1.4 | 14.9 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 1.3 | 6.7 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 1.3 | 6.6 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 1.3 | 3.9 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 1.3 | 1.3 | GO:0034418 | urate biosynthetic process(GO:0034418) |
| 1.2 | 4.9 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 1.2 | 3.6 | GO:0006574 | valine catabolic process(GO:0006574) |
| 1.2 | 4.6 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 1.2 | 10.4 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 1.1 | 27.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.0 | 7.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 1.0 | 3.0 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 1.0 | 5.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.0 | 4.0 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 1.0 | 3.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) regulation of Toll signaling pathway(GO:0008592) negative regulation of Toll signaling pathway(GO:0045751) |
| 1.0 | 1.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.8 | 3.4 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.8 | 3.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.8 | 25.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.8 | 2.3 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.8 | 3.9 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.7 | 2.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.7 | 4.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.7 | 3.0 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.7 | 4.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.7 | 17.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.7 | 9.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.6 | 1.9 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.6 | 1.9 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.6 | 2.5 | GO:0098705 | copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.5 | 8.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.5 | 2.2 | GO:0035772 | interleukin-13-mediated signaling pathway(GO:0035772) |
| 0.5 | 2.6 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.5 | 5.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.5 | 1.6 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.5 | 3.5 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.5 | 3.0 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.5 | 4.4 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.5 | 4.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.5 | 4.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.5 | 1.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.5 | 5.7 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.5 | 2.8 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.5 | 1.4 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.5 | 1.4 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.5 | 3.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.4 | 2.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 4.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.4 | 2.2 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.4 | 3.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.4 | 1.3 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.4 | 8.7 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.4 | 3.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.4 | 3.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.4 | 2.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.4 | 0.9 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.4 | 3.8 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.4 | 1.3 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.4 | 12.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.4 | 1.2 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) regulation of tau-protein kinase activity(GO:1902947) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.4 | 3.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 | 2.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.4 | 6.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.4 | 4.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.4 | 1.5 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.4 | 2.5 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.4 | 1.8 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.4 | 2.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.4 | 4.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.4 | 2.8 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.4 | 1.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.3 | 4.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.3 | 3.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 1.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.3 | 1.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 1.3 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.3 | 1.0 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.3 | 3.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 1.9 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.3 | 3.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 1.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.3 | 1.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.3 | 5.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.3 | 4.0 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.3 | 2.0 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.3 | 4.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.3 | 3.7 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.3 | 0.9 | GO:0019677 | NADP catabolic process(GO:0006742) NAD catabolic process(GO:0019677) |
| 0.3 | 1.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.3 | 0.8 | GO:0045963 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.3 | 4.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.3 | 2.3 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 | 1.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.3 | 0.8 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.3 | 1.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.3 | 2.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 1.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.3 | 1.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 3.1 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.3 | 1.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 1.3 | GO:0009758 | carbohydrate utilization(GO:0009758) |
| 0.3 | 0.5 | GO:0016107 | sesquiterpenoid metabolic process(GO:0006714) polyprenol catabolic process(GO:0016095) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.2 | 0.2 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.2 | 1.4 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 1.0 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.2 | 1.1 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 1.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 1.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 1.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 4.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 0.9 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.2 | 0.7 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 0.9 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 | 0.9 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.2 | 1.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 0.6 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 0.8 | GO:0019585 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) cellular glucuronidation(GO:0052695) |
| 0.2 | 8.6 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.2 | 0.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 6.7 | GO:0042311 | vasodilation(GO:0042311) |
| 0.2 | 3.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.2 | 0.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 2.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 1.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 2.6 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 0.5 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 1.2 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.2 | 3.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 2.4 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.2 | 0.5 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.2 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 0.7 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.2 | 3.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 4.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.7 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.2 | 0.8 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 0.7 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.2 | 0.6 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 4.9 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.2 | 2.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 1.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 9.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.9 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 1.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.3 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.1 | 2.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.4 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 0.7 | GO:0045356 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) positive regulation of chemokine biosynthetic process(GO:0045080) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.5 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 0.8 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 1.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 1.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.5 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 | 2.9 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.7 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.3 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.1 | 1.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.5 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 1.4 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 2.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 2.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.5 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 2.0 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.7 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 0.5 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.4 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 1.2 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.1 | 1.7 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.7 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.1 | 0.3 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.1 | 0.7 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.1 | 0.3 | GO:0042441 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 0.1 | 0.4 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.1 | 1.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 2.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.5 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.8 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.1 | 1.2 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.7 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.5 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 0.8 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.1 | 0.2 | GO:0000962 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.1 | 1.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.8 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 1.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.9 | GO:0032275 | luteinizing hormone secretion(GO:0032275) |
| 0.1 | 1.0 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.5 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.1 | 0.4 | GO:0060022 | hard palate development(GO:0060022) |
| 0.1 | 0.3 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.3 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 1.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.2 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 1.4 | GO:0060039 | pericardium development(GO:0060039) |
| 0.1 | 0.3 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 0.5 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.2 | GO:0043201 | response to leucine(GO:0043201) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 1.7 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.8 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.1 | 0.3 | GO:0031645 | negative regulation of neurological system process(GO:0031645) |
| 0.1 | 1.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.5 | GO:0030432 | urinary bladder smooth muscle contraction(GO:0014832) peristalsis(GO:0030432) |
| 0.1 | 1.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 1.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 | 0.7 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.1 | 0.7 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.1 | 0.7 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 0.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.4 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) cornification(GO:0070268) |
| 0.0 | 0.6 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 6.2 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.0 | 11.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.2 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 1.1 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 1.0 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.5 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.4 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 4.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 1.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0051177 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 1.3 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.5 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.6 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0070471 | uterine smooth muscle contraction(GO:0070471) regulation of uterine smooth muscle contraction(GO:0070472) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.0 | 0.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 1.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.6 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 | 0.5 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.8 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 28.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.3 | GO:0070972 | protein localization to endoplasmic reticulum(GO:0070972) |
| 0.0 | 0.6 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 1.7 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.0 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.2 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0046496 | pyridine nucleotide metabolic process(GO:0019362) nicotinamide nucleotide metabolic process(GO:0046496) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.2 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.0 | 0.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 5.6 | GO:0005975 | carbohydrate metabolic process(GO:0005975) |
| 0.0 | 0.2 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 32.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.4 | 16.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 1.3 | 6.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.3 | 5.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.3 | 16.4 | GO:0042627 | chylomicron(GO:0042627) |
| 1.2 | 3.5 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 1.0 | 8.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 9.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.7 | 12.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.7 | 12.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.7 | 76.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.7 | 14.1 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.6 | 1.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.5 | 10.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.5 | 10.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 10.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.4 | 1.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.4 | 9.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 1.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 1.0 | GO:0045203 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.3 | 4.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 2.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 1.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 7.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 1.6 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.3 | 4.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 1.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 2.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 1.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 5.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 1.9 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 2.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 11.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 1.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 1.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 3.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.2 | 1.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 1.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 2.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 3.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 0.5 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 23.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 11.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 3.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 104.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 0.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.2 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.1 | 1.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.7 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.4 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.1 | 2.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 6.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 5.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 22.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 7.6 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 0.6 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 10.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 50.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.6 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 3.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 9.9 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.0 | GO:0061574 | ASAP complex(GO:0061574) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.8 | 47.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 5.5 | 16.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 3.7 | 11.0 | GO:0052852 | very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 3.3 | 10.0 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 3.3 | 22.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 2.7 | 13.7 | GO:0005534 | galactose binding(GO:0005534) |
| 2.5 | 12.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 2.2 | 28.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 2.0 | 6.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 2.0 | 5.9 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 1.9 | 9.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 1.8 | 9.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 1.7 | 5.2 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 1.7 | 6.9 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 1.7 | 5.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 1.7 | 50.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.6 | 6.6 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 1.5 | 4.5 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 1.5 | 7.4 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 1.3 | 12.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 1.3 | 3.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.3 | 34.2 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.2 | 4.7 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 1.1 | 10.3 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 1.1 | 17.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 1.1 | 3.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 1.0 | 4.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.0 | 3.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.0 | 3.9 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.9 | 8.8 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.9 | 4.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.8 | 12.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.8 | 2.4 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.8 | 2.3 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.8 | 3.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.7 | 4.4 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.7 | 4.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.7 | 2.8 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.6 | 75.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.6 | 6.9 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.6 | 3.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.6 | 10.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.6 | 15.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.6 | 2.8 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.6 | 2.8 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.5 | 4.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.5 | 5.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.5 | 4.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 5.7 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.5 | 2.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.5 | 3.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.5 | 1.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.4 | 2.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 1.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.4 | 1.7 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.4 | 2.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.4 | 2.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.4 | 1.9 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.4 | 1.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.3 | 2.4 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 1.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 1.0 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 1.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.3 | 3.5 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.3 | 3.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 1.9 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.3 | 0.9 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.3 | 1.8 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 3.7 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.3 | 0.8 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.3 | 4.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 2.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 3.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 0.5 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.3 | 2.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.3 | 4.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 3.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.3 | 0.5 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.3 | 3.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 1.2 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.2 | 0.9 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 1.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 0.9 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 0.9 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.2 | 3.0 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 0.9 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 0.6 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.2 | 5.9 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.2 | 4.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.2 | 1.8 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 1.0 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 1.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 2.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 1.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 0.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 3.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 1.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 0.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.2 | 0.5 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 0.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 4.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 0.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 0.3 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.2 | 1.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.2 | 13.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 2.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 0.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 1.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 3.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.4 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 1.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 4.1 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.1 | 4.3 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.1 | 0.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 2.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.0 | GO:0016885 | CoA carboxylase activity(GO:0016421) ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.1 | 1.7 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.7 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 1.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 3.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 3.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.5 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.8 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 2.8 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.1 | 1.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.6 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 8.6 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 2.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.0 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.6 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 15.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 6.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 8.9 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.1 | 1.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.6 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 2.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 7.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.1 | GO:0017171 | serine hydrolase activity(GO:0017171) |
| 0.1 | 0.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.7 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.1 | 0.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 21.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 5.1 | GO:0016407 | acetyltransferase activity(GO:0016407) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 2.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 2.0 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 1.9 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 1.5 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.0 | 3.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.8 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 3.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0016410 | N-acyltransferase activity(GO:0016410) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 14.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 16.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 4.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 4.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 49.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 4.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 9.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 0.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 3.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 11.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 1.9 | 65.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.4 | 28.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.3 | 17.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.1 | 14.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 19.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.6 | 11.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.6 | 1.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.5 | 4.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 6.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 7.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 3.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 3.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 6.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 9.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 3.0 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.3 | 4.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.3 | 12.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 11.9 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.3 | 2.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.3 | 4.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 9.3 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.2 | 6.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 8.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 3.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 3.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 2.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 1.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 1.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 3.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 1.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 1.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 6.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 3.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 20.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 0.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.7 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 2.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.1 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |