Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
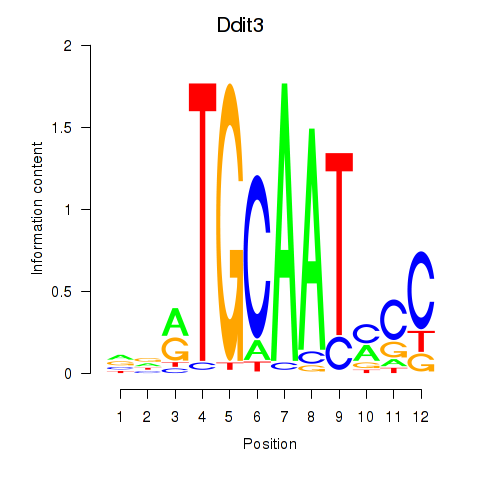
Results for Ddit3
Z-value: 0.86
Transcription factors associated with Ddit3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ddit3
|
ENSRNOG00000006789 | DNA-damage inducible transcript 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ddit3 | rn6_v1_chr7_+_70580198_70580252 | -0.11 | 5.3e-02 | Click! |
Activity profile of Ddit3 motif
Sorted Z-values of Ddit3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_86297623 | 28.79 |
ENSRNOT00000067162
ENSRNOT00000081607 ENSRNOT00000085265 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II beta |
| chr3_-_44086006 | 28.24 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr2_+_200793571 | 25.20 |
ENSRNOT00000091444
|
Hao2
|
hydroxyacid oxidase 2 |
| chr16_-_49820235 | 23.13 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr11_+_60073383 | 22.76 |
ENSRNOT00000087508
|
Tagln3
|
transgelin 3 |
| chr15_-_71779033 | 20.02 |
ENSRNOT00000017765
|
Pcdh20
|
protocadherin 20 |
| chr10_-_83898527 | 17.96 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chrX_+_118084890 | 16.51 |
ENSRNOT00000065293
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr18_+_57654290 | 14.94 |
ENSRNOT00000025892
|
Htr4
|
5-hydroxytryptamine receptor 4 |
| chr11_-_81717521 | 14.84 |
ENSRNOT00000058422
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chrX_+_39711201 | 14.60 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr19_+_50848736 | 13.81 |
ENSRNOT00000077053
|
Cdh13
|
cadherin 13 |
| chr8_+_39305128 | 13.75 |
ENSRNOT00000008285
|
Fez1
|
fasciculation and elongation protein zeta 1 |
| chr3_-_158328881 | 13.67 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr7_+_64672722 | 13.59 |
ENSRNOT00000064448
ENSRNOT00000005539 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr7_+_12433933 | 12.79 |
ENSRNOT00000060690
|
Cbarp
|
CACN beta subunit associated regulatory protein |
| chr18_+_30387937 | 12.61 |
ENSRNOT00000027210
|
Pcdhb4
|
protocadherin beta 4 |
| chr20_-_48503898 | 12.44 |
ENSRNOT00000073091
|
Wasf1
|
WAS protein family, member 1 |
| chr8_-_59139946 | 12.43 |
ENSRNOT00000078571
|
Cib2
|
calcium and integrin binding family member 2 |
| chr9_-_8632017 | 11.70 |
ENSRNOT00000043269
|
AABR07066416.1
|
|
| chr16_-_75309176 | 11.49 |
ENSRNOT00000018427
|
Defb1
|
defensin beta 1 |
| chr8_-_49106177 | 11.05 |
ENSRNOT00000067558
|
Tmem25
|
transmembrane protein 25 |
| chr10_+_37215937 | 11.00 |
ENSRNOT00000006567
|
Sar1b
|
secretion associated, Ras related GTPase 1B |
| chr15_+_17834635 | 10.82 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr12_+_32103198 | 10.76 |
ENSRNOT00000085464
|
Tmem132d
|
transmembrane protein 132D |
| chr18_+_30527705 | 10.70 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr4_-_86466797 | 10.61 |
ENSRNOT00000017531
|
Pde1c
|
phosphodiesterase 1C |
| chr11_-_35098883 | 10.59 |
ENSRNOT00000079955
|
Kcnj6
|
potassium voltage-gated channel subfamily J member 6 |
| chr7_-_82687130 | 10.57 |
ENSRNOT00000006791
|
Tmem74
|
transmembrane protein 74 |
| chr7_+_80625010 | 10.45 |
ENSRNOT00000084940
|
Oxr1
|
oxidation resistance 1 |
| chr9_-_4945352 | 10.40 |
ENSRNOT00000082530
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr3_+_111160205 | 9.95 |
ENSRNOT00000019392
|
Chac1
|
ChaC glutathione-specific gamma-glutamylcyclotransferase 1 |
| chr14_-_82048251 | 9.83 |
ENSRNOT00000074734
|
Nat8l
|
N-acetyltransferase 8-like |
| chr1_-_233145924 | 9.81 |
ENSRNOT00000018763
ENSRNOT00000082442 |
Psat1
|
phosphoserine aminotransferase 1 |
| chr3_-_45210474 | 9.69 |
ENSRNOT00000091777
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr1_-_215033460 | 9.64 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr9_-_9142339 | 9.45 |
ENSRNOT00000046852
|
MGC116197
|
similar to RIKEN cDNA 1700001E04 |
| chr6_-_1319541 | 9.35 |
ENSRNOT00000006527
|
Strn
|
striatin |
| chr2_-_53185926 | 9.29 |
ENSRNOT00000081558
|
Ghr
|
growth hormone receptor |
| chr10_-_104748003 | 9.27 |
ENSRNOT00000042372
ENSRNOT00000046754 |
Acox1
|
acyl-CoA oxidase 1 |
| chr19_-_6031655 | 9.27 |
ENSRNOT00000092016
|
AABR07042733.2
|
|
| chr9_-_30844199 | 9.22 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr18_+_31444472 | 9.05 |
ENSRNOT00000075159
|
Rnf14
|
ring finger protein 14 |
| chr19_+_24044103 | 9.04 |
ENSRNOT00000004621
|
Rnf150
|
ring finger protein 150 |
| chr3_+_147226004 | 8.84 |
ENSRNOT00000012765
|
Tmem74b
|
transmembrane protein 74B |
| chr1_+_100131449 | 8.69 |
ENSRNOT00000091964
ENSRNOT00000071416 |
Klk1
|
kallikrein 1 |
| chr12_-_2592838 | 8.53 |
ENSRNOT00000079918
|
Evi5l
|
ecotropic viral integration site 5-like |
| chr9_-_26707571 | 8.20 |
ENSRNOT00000080948
|
AABR07067023.1
|
|
| chrX_+_33884499 | 7.99 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr1_-_222350173 | 7.97 |
ENSRNOT00000030625
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr16_+_23668595 | 7.64 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr4_+_34962339 | 7.59 |
ENSRNOT00000086248
|
AABR07059778.1
|
|
| chr10_+_40543288 | 7.18 |
ENSRNOT00000016755
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr14_-_42560174 | 7.05 |
ENSRNOT00000003128
|
Tmem33
|
transmembrane protein 33 |
| chr1_+_157573324 | 6.99 |
ENSRNOT00000092066
|
Rab30
|
RAB30, member RAS oncogene family |
| chr1_+_48273611 | 6.94 |
ENSRNOT00000022254
ENSRNOT00000022068 |
Slc22a1
|
solute carrier family 22 member 1 |
| chr20_-_48881119 | 6.73 |
ENSRNOT00000018892
|
Qrsl1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr2_+_116970344 | 6.33 |
ENSRNOT00000039603
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr20_-_5123073 | 6.27 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chr2_-_148722263 | 6.27 |
ENSRNOT00000017868
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chrM_-_14061 | 6.10 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr3_-_143899370 | 6.05 |
ENSRNOT00000049635
|
Andpro
|
androgen regulated protein |
| chr11_-_61874327 | 5.96 |
ENSRNOT00000089057
|
Drd3
|
dopamine receptor D3 |
| chr2_-_216382244 | 5.89 |
ENSRNOT00000086695
ENSRNOT00000087259 |
LOC103689940
|
pancreatic alpha-amylase-like |
| chr1_-_166943592 | 5.79 |
ENSRNOT00000026962
|
Folr1
|
folate receptor 1 |
| chr11_+_9642365 | 5.79 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr4_+_120672152 | 5.76 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr9_-_99680979 | 5.74 |
ENSRNOT00000049347
|
Olr1345
|
olfactory receptor 1345 |
| chr8_-_21831668 | 5.71 |
ENSRNOT00000027897
|
Col5a3
|
collagen type V alpha 3 chain |
| chr10_+_44547564 | 5.61 |
ENSRNOT00000047914
|
Olr1443
|
olfactory receptor 1443 |
| chr5_+_33580944 | 5.51 |
ENSRNOT00000092054
ENSRNOT00000036050 |
Rmdn1
|
regulator of microtubule dynamics 1 |
| chr6_-_26241337 | 5.50 |
ENSRNOT00000006525
|
Slc4a1ap
|
solute carrier family 4 member 1 adaptor protein |
| chr10_-_81653717 | 5.45 |
ENSRNOT00000003611
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr10_+_98706960 | 5.36 |
ENSRNOT00000006217
|
Map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr5_+_14415841 | 5.36 |
ENSRNOT00000010682
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr11_-_77703255 | 4.89 |
ENSRNOT00000083319
|
Cldn16
|
claudin 16 |
| chr19_+_24670699 | 4.86 |
ENSRNOT00000038213
|
Olr1667
|
olfactory receptor 1667 |
| chr2_+_85377318 | 4.83 |
ENSRNOT00000016506
ENSRNOT00000085094 |
Sema5a
|
semaphorin 5A |
| chr1_+_7658890 | 4.76 |
ENSRNOT00000020946
|
Fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr1_-_167588630 | 4.71 |
ENSRNOT00000050707
|
Olr39
|
olfactory receptor 39 |
| chr12_-_47793534 | 4.58 |
ENSRNOT00000001588
|
Fam222a
|
family with sequence similarity 222, member A |
| chr12_-_8156151 | 4.43 |
ENSRNOT00000064721
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr8_+_116297950 | 4.32 |
ENSRNOT00000051522
|
Nprl2
|
NPR2-like, GATOR1 complex subunit |
| chr6_-_28994519 | 4.32 |
ENSRNOT00000006668
|
Ubxn2a
|
UBX domain protein 2A |
| chr15_+_61069581 | 4.29 |
ENSRNOT00000084333
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr2_-_216348194 | 4.23 |
ENSRNOT00000087839
|
LOC103689940
|
pancreatic alpha-amylase-like |
| chr8_+_82037977 | 4.22 |
ENSRNOT00000082288
|
Myo5a
|
myosin VA |
| chr1_-_173469699 | 4.15 |
ENSRNOT00000040345
|
Olr281
|
olfactory receptor 281 |
| chr10_-_45514878 | 4.10 |
ENSRNOT00000036940
|
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr20_+_48881194 | 4.00 |
ENSRNOT00000000304
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr1_+_267618248 | 3.97 |
ENSRNOT00000017186
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr1_+_41590494 | 3.92 |
ENSRNOT00000089984
|
Esr1
|
estrogen receptor 1 |
| chr19_-_11057254 | 3.84 |
ENSRNOT00000025559
|
Herpud1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr10_-_37215899 | 3.68 |
ENSRNOT00000006356
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr1_+_174655939 | 3.63 |
ENSRNOT00000014002
|
Ipo7
|
importin 7 |
| chr8_+_43872728 | 3.48 |
ENSRNOT00000079702
|
Olr1337
|
olfactory receptor 1337 |
| chr1_-_174379566 | 3.47 |
ENSRNOT00000067388
|
Tmem9b
|
TMEM9 domain family, member B |
| chr6_+_26241672 | 3.45 |
ENSRNOT00000006543
|
Supt7l
|
SPT7-like STAGA complex gamma subunit |
| chrX_-_105568343 | 3.39 |
ENSRNOT00000029807
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr1_-_264294630 | 3.36 |
ENSRNOT00000035758
|
Sec31b
|
SEC31 homolog B, COPII coat complex component |
| chr1_-_229804614 | 3.17 |
ENSRNOT00000049665
|
Olr343
|
olfactory receptor 343 |
| chr1_-_228524439 | 3.06 |
ENSRNOT00000028601
|
Olr323
|
olfactory receptor 323 |
| chrX_-_33722090 | 2.92 |
ENSRNOT00000059010
|
Rpl39
|
ribosomal protein L39 |
| chr11_+_80319177 | 2.83 |
ENSRNOT00000036777
|
Rtp2
|
receptor (chemosensory) transporter protein 2 |
| chr5_-_164927869 | 2.78 |
ENSRNOT00000012080
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr10_-_95431312 | 2.77 |
ENSRNOT00000085552
|
AABR07030603.2
|
|
| chr2_-_122690617 | 2.75 |
ENSRNOT00000018942
|
Mccc1
|
methylcrotonoyl-CoA carboxylase 1 |
| chr19_+_26194465 | 2.74 |
ENSRNOT00000087627
ENSRNOT00000065931 |
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr3_-_77661863 | 2.70 |
ENSRNOT00000080807
|
Olr668
|
olfactory receptor 668 |
| chr18_+_53088994 | 2.69 |
ENSRNOT00000091921
|
AC104053.2
|
|
| chr3_+_102925471 | 2.64 |
ENSRNOT00000040364
|
Olr772
|
olfactory receptor 772 |
| chr7_-_124367630 | 2.64 |
ENSRNOT00000055978
|
Ttll1
|
tubulin tyrosine ligase like 1 |
| chr1_+_72356880 | 2.49 |
ENSRNOT00000021917
|
Zfp865
|
zinc finger protein 865 |
| chr1_-_214067657 | 2.46 |
ENSRNOT00000044390
|
LOC100911881
|
chitinase domain-containing protein 1-like |
| chr4_+_66276835 | 2.46 |
ENSRNOT00000007544
|
Fmc1
|
formation of mitochondrial complex V assembly factor 1 |
| chr4_+_22082194 | 2.42 |
ENSRNOT00000091799
|
Crot
|
carnitine O-octanoyltransferase |
| chr5_+_106952082 | 2.41 |
ENSRNOT00000046931
|
RGD1561246
|
similar to put. precursor MuIFN-alpha 5 |
| chr14_-_10749120 | 2.36 |
ENSRNOT00000003046
|
Cops4
|
COP9 signalosome subunit 4 |
| chr5_-_137874692 | 2.36 |
ENSRNOT00000089549
|
Olr867
|
olfactory receptor 867 |
| chr14_+_104250617 | 2.36 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr2_+_181987217 | 2.35 |
ENSRNOT00000034521
|
Fgg
|
fibrinogen gamma chain |
| chr12_+_8032809 | 2.34 |
ENSRNOT00000082850
|
Slc7a1
|
solute carrier family 7 member 1 |
| chr3_-_143807287 | 2.29 |
ENSRNOT00000046355
ENSRNOT00000081843 |
AABR07054186.1
|
|
| chr1_-_173129038 | 2.25 |
ENSRNOT00000075693
|
LOC100912408
|
olfactory receptor 10A3-like |
| chr10_-_62366044 | 2.21 |
ENSRNOT00000045673
|
Olr1474
|
olfactory receptor 1474 |
| chr6_-_111131271 | 2.14 |
ENSRNOT00000081727
|
Ngb
|
neuroglobin |
| chr7_-_140502441 | 2.10 |
ENSRNOT00000089544
|
Kmt2d
|
lysine methyltransferase 2D |
| chr6_-_23581052 | 2.06 |
ENSRNOT00000006190
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr8_-_116297850 | 2.03 |
ENSRNOT00000031739
ENSRNOT00000082320 |
Cyb561d2
|
cytochrome b561 family, member D2 |
| chr10_+_96661696 | 1.98 |
ENSRNOT00000077567
|
AABR07030630.1
|
|
| chr14_-_37361798 | 1.96 |
ENSRNOT00000002980
|
Cwh43
|
cell wall biogenesis 43 C-terminal homolog |
| chr1_-_214504569 | 1.90 |
ENSRNOT00000082221
|
Chid1
|
chitinase domain containing 1 |
| chrX_-_119162518 | 1.89 |
ENSRNOT00000073176
|
LOC103694506
|
ubiquitin-conjugating enzyme E2 W |
| chr2_+_93758919 | 1.89 |
ENSRNOT00000077782
|
Fabp12
|
fatty acid binding protein 12 |
| chr2_-_96032722 | 1.87 |
ENSRNOT00000015746
|
LOC100360846
|
proteasome subunit beta type 6-like |
| chr15_+_33004133 | 1.81 |
ENSRNOT00000013246
|
Oxa1l
|
OXA1L, mitochondrial inner membrane protein |
| chr5_+_27326762 | 1.69 |
ENSRNOT00000066191
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr2_-_260108897 | 1.68 |
ENSRNOT00000014106
|
Msh4
|
mutS homolog 4 |
| chr7_+_16304954 | 1.61 |
ENSRNOT00000050751
|
Olr927
|
olfactory receptor 927 |
| chr2_-_38110567 | 1.56 |
ENSRNOT00000072212
|
Ipo11
|
importin 11 |
| chr4_+_174692331 | 1.54 |
ENSRNOT00000011770
|
Plekha5
|
pleckstrin homology domain containing A5 |
| chr15_+_39638510 | 1.53 |
ENSRNOT00000037800
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr7_+_9262982 | 1.49 |
ENSRNOT00000047647
|
Olr1063
|
olfactory receptor 1063 |
| chr1_-_230766515 | 1.44 |
ENSRNOT00000040049
|
Olr383
|
olfactory receptor 383 |
| chr1_+_101397828 | 1.42 |
ENSRNOT00000028189
|
Kcna7
|
potassium voltage-gated channel subfamily A member 7 |
| chr12_+_17253791 | 1.39 |
ENSRNOT00000083814
|
Zfand2a
|
zinc finger AN1-type containing 2A |
| chr20_+_13965121 | 1.37 |
ENSRNOT00000089005
ENSRNOT00000066773 ENSRNOT00000079996 |
Susd2
|
sushi domain containing 2 |
| chr10_+_105073077 | 1.32 |
ENSRNOT00000083022
ENSRNOT00000086382 ENSRNOT00000087084 |
Ten1
|
TEN1 CST complex subunit |
| chr1_+_218058405 | 1.29 |
ENSRNOT00000028365
|
Fgf19
|
fibroblast growth factor 19 |
| chr17_+_6684621 | 1.26 |
ENSRNOT00000089138
|
RGD1311345
|
similar to CG9752-PA |
| chr9_-_99659425 | 1.23 |
ENSRNOT00000051686
|
Olr1343
|
olfactory receptor 1343 |
| chr15_-_36213666 | 1.19 |
ENSRNOT00000078704
|
Olr1291
|
olfactory receptor 1291 |
| chr9_-_16406338 | 1.18 |
ENSRNOT00000073079
|
Tbcc
|
tubulin folding cofactor C |
| chr20_+_1110773 | 1.17 |
ENSRNOT00000041697
|
Olr1702
|
olfactory receptor 1702 |
| chr7_+_70335061 | 1.15 |
ENSRNOT00000072176
ENSRNOT00000084933 |
Cyp27b1
|
cytochrome P450, family 27, subfamily b, polypeptide 1 |
| chr2_-_112368557 | 1.10 |
ENSRNOT00000075017
|
AABR07009787.1
|
|
| chr1_-_71173216 | 1.09 |
ENSRNOT00000020523
|
Zfp28
|
zinc finger protein 28 |
| chr1_-_172943853 | 1.09 |
ENSRNOT00000047040
|
Olr278
|
olfactory receptor 278 |
| chr17_+_48436720 | 1.05 |
ENSRNOT00000074309
|
AABR07027902.1
|
|
| chr1_+_254531659 | 1.04 |
ENSRNOT00000091377
|
AC080157.1
|
|
| chr14_-_21771567 | 1.02 |
ENSRNOT00000038393
|
AABR07014573.1
|
|
| chr1_+_168668707 | 0.96 |
ENSRNOT00000015108
|
AC107531.3
|
|
| chr10_+_71536533 | 0.93 |
ENSRNOT00000088138
|
Acaca
|
acetyl-CoA carboxylase alpha |
| chr15_+_64939527 | 0.88 |
ENSRNOT00000045865
|
AABR07018556.1
|
|
| chr3_-_72219246 | 0.88 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr5_-_39689824 | 0.88 |
ENSRNOT00000046713
|
AABR07047606.1
|
|
| chr7_-_99808612 | 0.87 |
ENSRNOT00000074953
|
AABR07058091.2
|
|
| chr13_+_106751625 | 0.87 |
ENSRNOT00000004992
|
Ush2a
|
usherin |
| chr7_-_122403667 | 0.82 |
ENSRNOT00000088814
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr5_-_137846436 | 0.81 |
ENSRNOT00000046941
|
Olr866
|
olfactory receptor 866 |
| chrX_+_137934484 | 0.79 |
ENSRNOT00000049046
|
LOC317588
|
hypothetical protein LOC317588 |
| chr1_-_168869420 | 0.78 |
ENSRNOT00000021436
|
Olr125
|
olfactory receptor 125 |
| chr1_+_142679345 | 0.76 |
ENSRNOT00000034267
|
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr1_-_4560789 | 0.75 |
ENSRNOT00000082415
|
Adgb
|
androglobin |
| chr8_-_43055175 | 0.72 |
ENSRNOT00000074413
|
LOC103693054
|
olfactory receptor 8G1-like |
| chrX_+_70408064 | 0.71 |
ENSRNOT00000038924
|
Awat1
|
acyl-CoA wax alcohol acyltransferase 1 |
| chr3_+_75354237 | 0.70 |
ENSRNOT00000050634
|
Olr557
|
olfactory receptor 557 |
| chr4_-_155740193 | 0.63 |
ENSRNOT00000043229
|
LOC102550396
|
LRRGT00188 |
| chr5_-_172078760 | 0.61 |
ENSRNOT00000016781
|
Actrt2
|
actin-related protein T2 |
| chr1_+_207508414 | 0.60 |
ENSRNOT00000054896
|
Nps
|
neuropeptide S |
| chr4_-_166803127 | 0.60 |
ENSRNOT00000067909
|
RGD1306750
|
LOC362451 |
| chr1_+_169590308 | 0.60 |
ENSRNOT00000023147
|
Olr155
|
olfactory receptor 155 |
| chr1_+_79573118 | 0.53 |
ENSRNOT00000072039
|
Igfl3
|
IGF-like family member 3 |
| chr10_+_59923201 | 0.53 |
ENSRNOT00000073603
ENSRNOT00000045573 |
Olr1498
|
olfactory receptor 1498 |
| chr1_+_75326217 | 0.50 |
ENSRNOT00000044964
|
Vom1r62
|
vomeronasal 1 receptor 62 |
| chr1_+_190462327 | 0.50 |
ENSRNOT00000030732
|
LOC691519
|
similar to ankyrin repeat domain 26 |
| chr1_+_170796271 | 0.50 |
ENSRNOT00000042820
|
Olr210
|
olfactory receptor 210 |
| chr3_-_76718684 | 0.48 |
ENSRNOT00000050173
|
Olr631
|
olfactory receptor 631 |
| chr8_+_82038967 | 0.45 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr6_-_125812517 | 0.44 |
ENSRNOT00000007061
|
Trip11
|
thyroid hormone receptor interactor 11 |
| chr20_+_572454 | 0.43 |
ENSRNOT00000061839
|
Olr1684
|
olfactory receptor 1684 |
| chr1_+_88586837 | 0.42 |
ENSRNOT00000080555
|
Zfp382
|
zinc finger protein 382 |
| chr15_-_43542939 | 0.42 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr7_+_25919867 | 0.36 |
ENSRNOT00000009625
ENSRNOT00000090153 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr1_+_170887363 | 0.36 |
ENSRNOT00000036261
|
Olr215
|
olfactory receptor 215 |
| chr2_-_221099738 | 0.35 |
ENSRNOT00000022947
|
Snx7
|
sorting nexin 7 |
| chr11_-_87485833 | 0.31 |
ENSRNOT00000079948
|
AABR07034751.1
|
|
| chr3_+_16311578 | 0.27 |
ENSRNOT00000010341
|
Olr409
|
olfactory receptor 409 |
| chr1_-_171979933 | 0.24 |
ENSRNOT00000026744
|
Cyb5r2
|
cytochrome b5 reductase 2 |
| chr10_+_34672726 | 0.17 |
ENSRNOT00000043058
|
Olr1394
|
olfactory receptor 1394 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ddit3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 28.8 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 8.4 | 25.2 | GO:0018924 | mandelate metabolic process(GO:0018924) |
| 5.5 | 16.5 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 3.1 | 9.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 3.0 | 6.0 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 2.9 | 11.5 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 2.5 | 9.8 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 2.2 | 6.7 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 2.1 | 6.3 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 2.0 | 9.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.9 | 5.8 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 1.8 | 12.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 1.8 | 7.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 1.7 | 13.8 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 1.4 | 5.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.2 | 9.9 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 1.2 | 4.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 1.2 | 4.7 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.1 | 13.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 1.1 | 12.4 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 1.1 | 3.2 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 1.0 | 10.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 1.0 | 9.3 | GO:0046449 | allantoin metabolic process(GO:0000255) isoleucine metabolic process(GO:0006549) creatine metabolic process(GO:0006600) creatinine metabolic process(GO:0046449) |
| 1.0 | 3.9 | GO:1990375 | baculum development(GO:1990375) |
| 1.0 | 10.6 | GO:0071315 | cellular response to morphine(GO:0071315) |
| 0.9 | 2.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.9 | 7.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.9 | 3.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.9 | 12.8 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.8 | 10.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.8 | 5.4 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.8 | 3.8 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.7 | 4.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.7 | 6.9 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.7 | 8.5 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.6 | 14.9 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.6 | 18.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.6 | 14.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.6 | 4.8 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.6 | 1.8 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.6 | 1.1 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) |
| 0.5 | 19.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.5 | 2.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.5 | 3.7 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.4 | 12.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.4 | 4.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.4 | 2.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.4 | 6.3 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.3 | 4.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 2.3 | GO:1990822 | arginine transmembrane transport(GO:1903826) basic amino acid transmembrane transport(GO:1990822) |
| 0.3 | 7.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 4.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.3 | 9.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.3 | 0.8 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.3 | 2.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 1.3 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.2 | 34.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 0.9 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 1.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 6.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.2 | 3.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.2 | 2.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 2.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 2.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 4.1 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 2.9 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 28.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.2 | 1.9 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.2 | 2.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 8.0 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 2.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 10.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.8 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.1 | 0.9 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.6 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.1 | 0.8 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 6.5 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.1 | 2.8 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.1 | 1.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 1.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 1.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 9.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 12.1 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.8 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 49.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 10.4 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.0 | 0.4 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 4.6 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 1.4 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) |
| 0.0 | 0.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 8.6 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 28.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 2.5 | 12.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 2.2 | 6.7 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 1.9 | 9.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.6 | 4.7 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.4 | 12.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 1.4 | 18.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.3 | 3.8 | GO:1990037 | Lewy body core(GO:1990037) |
| 1.3 | 6.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.9 | 4.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.8 | 28.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) spindle midzone(GO:0051233) |
| 0.6 | 1.7 | GO:0005713 | recombination nodule(GO:0005713) MutSalpha complex(GO:0032301) |
| 0.4 | 18.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.4 | 11.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.4 | 1.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.4 | 3.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 9.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 5.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 9.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 2.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 5.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 3.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.3 | 3.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 11.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 35.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 2.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 5.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 11.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 2.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 51.3 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 0.9 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 27.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 7.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 4.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 3.4 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.1 | 16.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 8.0 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 6.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 8.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 2.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 19.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 1.8 | GO:0098573 | integral component of mitochondrial membrane(GO:0032592) intrinsic component of mitochondrial membrane(GO:0098573) |
| 0.1 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 4.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 50.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 8.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 2.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 4.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 9.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 9.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 6.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 117.7 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 25.2 | GO:0052854 | very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 5.5 | 16.5 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 3.7 | 14.8 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 3.5 | 13.8 | GO:0055100 | adiponectin binding(GO:0055100) |
| 3.4 | 10.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 2.9 | 11.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 2.7 | 10.6 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 2.6 | 10.4 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 2.5 | 9.9 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 2.4 | 7.2 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 2.3 | 9.3 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 2.3 | 6.9 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 2.2 | 6.7 | GO:0050567 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 2.1 | 10.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.9 | 9.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 1.4 | 5.8 | GO:0051870 | methotrexate binding(GO:0051870) |
| 1.3 | 28.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 1.1 | 13.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.0 | 4.0 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.9 | 9.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.9 | 18.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.9 | 10.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.9 | 14.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.8 | 2.4 | GO:0008458 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 0.8 | 3.9 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.7 | 5.4 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.7 | 13.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.7 | 6.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.6 | 9.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.6 | 2.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.6 | 2.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.5 | 3.7 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.4 | 5.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 13.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.4 | 7.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.4 | 9.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.3 | 2.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 4.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.3 | 3.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 7.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 2.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.3 | 9.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 4.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 12.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 9.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 6.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 41.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 4.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 2.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 3.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 16.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 10.5 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 6.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 18.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 12.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 4.8 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 8.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 49.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 11.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 2.2 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 1.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.7 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 7.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 28.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 12.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 13.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 14.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.3 | 13.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 5.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 9.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 3.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 4.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 5.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 4.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 2.2 | 31.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.6 | 28.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 1.1 | 18.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.8 | 6.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.6 | 11.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 6.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.5 | 9.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.5 | 7.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.5 | 13.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 9.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.4 | 10.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 5.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 4.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 5.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 9.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 6.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 4.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 5.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 2.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 4.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 5.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 3.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 2.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 4.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 6.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |