Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
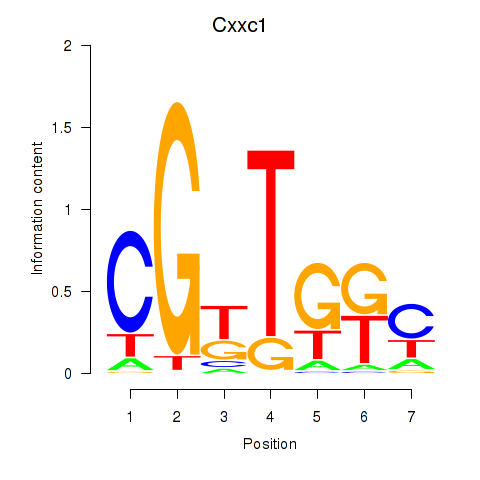
Results for Cxxc1
Z-value: 0.18
Transcription factors associated with Cxxc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cxxc1
|
ENSRNOG00000014614 | CXXC finger protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cxxc1 | rn6_v1_chr18_+_70192493_70192493 | -0.07 | 2.2e-01 | Click! |
Activity profile of Cxxc1 motif
Sorted Z-values of Cxxc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_257614731 | 2.69 |
ENSRNOT00000075680
|
LOC100911660
|
lymphocyte-specific helicase-like |
| chr1_+_257615088 | 2.25 |
ENSRNOT00000083183
|
LOC100911660
|
lymphocyte-specific helicase-like |
| chr10_-_57653359 | 1.64 |
ENSRNOT00000089638
|
Derl2
|
derlin 2 |
| chr1_+_234252757 | 1.56 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr4_+_85009350 | 1.22 |
ENSRNOT00000072271
|
Znrf2
|
zinc and ring finger 2 |
| chr1_-_101664436 | 0.99 |
ENSRNOT00000028522
|
Sec1
|
secretory blood group 1 |
| chr12_-_52506487 | 0.97 |
ENSRNOT00000089449
|
Ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr1_-_15374850 | 0.89 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr4_+_99823252 | 0.89 |
ENSRNOT00000013587
|
Polr1a
|
RNA polymerase I subunit A |
| chr1_-_101883744 | 0.84 |
ENSRNOT00000028635
|
Syngr4
|
synaptogyrin 4 |
| chr7_+_12006710 | 0.69 |
ENSRNOT00000045421
|
Klf16
|
Kruppel-like factor 16 |
| chr20_-_46666830 | 0.59 |
ENSRNOT00000000331
|
Cep57l1
|
centrosomal protein 57-like 1 |
| chr9_+_66058047 | 0.56 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr3_-_153188915 | 0.52 |
ENSRNOT00000079893
|
Soga1
|
suppressor of glucose, autophagy associated 1 |
| chr6_+_33786197 | 0.38 |
ENSRNOT00000082598
|
Pum2
|
pumilio RNA-binding family member 2 |
| chr1_+_101884276 | 0.26 |
ENSRNOT00000082917
|
Tmem143
|
transmembrane protein 143 |
| chr2_+_148722231 | 0.26 |
ENSRNOT00000018022
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr1_+_101884019 | 0.24 |
ENSRNOT00000028650
|
Tmem143
|
transmembrane protein 143 |
| chr12_+_36638457 | 0.21 |
ENSRNOT00000085692
|
Ubc
|
ubiquitin C |
| chr17_-_9558624 | 0.17 |
ENSRNOT00000036223
ENSRNOT00000083492 |
B4galt7
|
beta-1,4-galactosyltransferase 7 |
| chr4_+_26470864 | 0.16 |
ENSRNOT00000021979
|
Fzd1
|
frizzled class receptor 1 |
| chr6_+_136041777 | 0.15 |
ENSRNOT00000014395
|
Mark3
|
microtubule affinity regulating kinase 3 |
| chr8_+_85413537 | 0.13 |
ENSRNOT00000078159
|
Ick
|
intestinal cell kinase |
| chr6_+_136042059 | 0.08 |
ENSRNOT00000088784
|
Mark3
|
microtubule affinity regulating kinase 3 |
| chr5_+_135735825 | 0.00 |
ENSRNOT00000068267
|
Zswim5
|
zinc finger, SWIM-type containing 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cxxc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.6 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 1.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) retinal rod cell development(GO:0046548) retinal cone cell development(GO:0046549) |
| 0.1 | 0.9 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.9 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.1 | 1.0 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.4 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.3 | 1.6 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.9 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 1.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |