Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
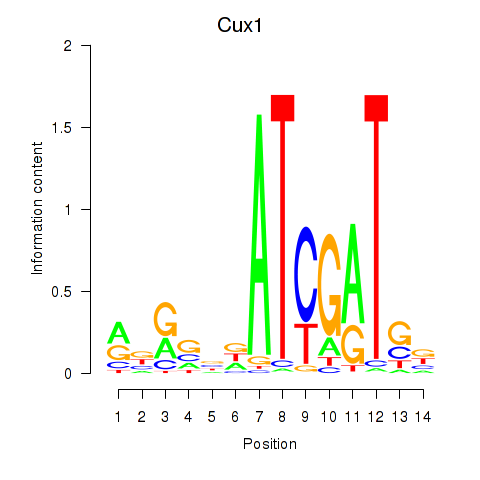
Results for Cux1
Z-value: 1.76
Transcription factors associated with Cux1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cux1
|
ENSRNOG00000001424 | cut-like homeobox 1 |
|
Cux1
|
ENSRNOG00000059116 | cut-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cux1 | rn6_v1_chr12_+_23151180_23151180 | -0.75 | 4.6e-58 | Click! |
Activity profile of Cux1 motif
Sorted Z-values of Cux1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_83744238 | 73.62 |
ENSRNOT00000028249
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr1_-_148119857 | 73.40 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr14_+_22375955 | 64.22 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr17_+_9736786 | 63.05 |
ENSRNOT00000081920
|
F12
|
coagulation factor XII |
| chr5_-_78985990 | 57.22 |
ENSRNOT00000009248
|
Ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr14_+_20266891 | 55.97 |
ENSRNOT00000004174
|
Gc
|
group specific component |
| chr1_+_83711251 | 54.26 |
ENSRNOT00000028237
ENSRNOT00000092008 |
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr2_+_23289374 | 53.46 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr4_-_161850875 | 53.17 |
ENSRNOT00000009467
|
Pzp
|
pregnancy-zone protein |
| chr15_+_18451144 | 51.56 |
ENSRNOT00000010260
|
Acox2
|
acyl-CoA oxidase 2 |
| chr9_-_4945352 | 51.45 |
ENSRNOT00000082530
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr4_-_115332052 | 50.04 |
ENSRNOT00000017643
|
Clec4f
|
C-type lectin domain family 4, member F |
| chr17_+_9736577 | 46.18 |
ENSRNOT00000066586
|
F12
|
coagulation factor XII |
| chr10_+_65767053 | 45.18 |
ENSRNOT00000078897
|
Vtn
|
vitronectin |
| chr17_-_69711689 | 44.72 |
ENSRNOT00000041925
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr13_-_47397890 | 44.47 |
ENSRNOT00000005505
|
C4bpb
|
complement component 4 binding protein, beta |
| chr19_-_50220455 | 44.18 |
ENSRNOT00000079760
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr6_-_127534247 | 43.11 |
ENSRNOT00000012500
|
Serpina6
|
serpin family A member 6 |
| chr7_-_71139267 | 41.48 |
ENSRNOT00000065232
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr1_+_147713892 | 41.10 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr4_-_50200328 | 40.19 |
ENSRNOT00000060530
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr7_-_129970550 | 37.84 |
ENSRNOT00000055879
|
Mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr6_-_127620296 | 37.46 |
ENSRNOT00000012577
|
Serpina1
|
serpin family A member 1 |
| chr2_-_180914940 | 37.38 |
ENSRNOT00000015732
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr16_+_18690649 | 37.14 |
ENSRNOT00000015190
|
Mat1a
|
methionine adenosyltransferase 1A |
| chrX_+_143097525 | 35.55 |
ENSRNOT00000004559
|
F9
|
coagulation factor IX |
| chr7_-_27552078 | 35.23 |
ENSRNOT00000059538
|
Stab2
|
stabilin 2 |
| chr5_+_124300477 | 34.46 |
ENSRNOT00000010100
|
C8b
|
complement C8 beta chain |
| chr5_-_124403195 | 33.94 |
ENSRNOT00000067850
|
C8a
|
complement C8 alpha chain |
| chr17_-_69404323 | 33.86 |
ENSRNOT00000051342
ENSRNOT00000066282 |
Akr1c2
|
aldo-keto reductase family 1, member C2 |
| chr5_+_134492756 | 33.83 |
ENSRNOT00000012888
ENSRNOT00000057095 ENSRNOT00000051385 |
Cyp4a1
|
cytochrome P450, family 4, subfamily a, polypeptide 1 |
| chr14_+_22724399 | 32.78 |
ENSRNOT00000002724
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr6_+_128050250 | 32.02 |
ENSRNOT00000077517
ENSRNOT00000013961 |
LOC500712
|
Ab1-233 |
| chr14_+_22553650 | 31.51 |
ENSRNOT00000092201
ENSRNOT00000002712 |
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr1_-_258877045 | 31.36 |
ENSRNOT00000071633
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr4_-_123557501 | 30.99 |
ENSRNOT00000075042
ENSRNOT00000085966 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr8_-_50526843 | 29.95 |
ENSRNOT00000092188
|
AABR07070085.1
|
|
| chr2_+_55775274 | 29.73 |
ENSRNOT00000018545
|
C9
|
complement C9 |
| chr7_+_34326087 | 29.66 |
ENSRNOT00000006971
|
Hal
|
histidine ammonia lyase |
| chr9_+_100281339 | 29.33 |
ENSRNOT00000029127
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr15_+_57290849 | 29.16 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chr17_-_43584152 | 29.14 |
ENSRNOT00000023241
|
Slc17a2
|
solute carrier family 17, member 2 |
| chr9_-_4978892 | 28.38 |
ENSRNOT00000015189
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr9_+_6970507 | 28.27 |
ENSRNOT00000079488
|
St6gal2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
| chr2_+_114413410 | 26.99 |
ENSRNOT00000015866
|
Slc2a2
|
solute carrier family 2 member 2 |
| chr8_-_50539331 | 26.67 |
ENSRNOT00000088997
|
AABR07073400.1
|
|
| chr2_+_20857202 | 26.57 |
ENSRNOT00000078919
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr13_+_51534025 | 26.14 |
ENSRNOT00000006637
|
Syt2
|
synaptotagmin 2 |
| chr1_+_213577122 | 25.40 |
ENSRNOT00000071925
|
RGD1309350
|
similar to transthyretin (4L369) |
| chr3_+_56861396 | 25.36 |
ENSRNOT00000000008
ENSRNOT00000084375 |
Gad1
|
glutamate decarboxylase 1 |
| chr10_+_66942398 | 25.33 |
ENSRNOT00000018986
|
Rab11fip4
|
RAB11 family interacting protein 4 |
| chr1_+_189364288 | 25.27 |
ENSRNOT00000080338
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr1_-_221431713 | 25.07 |
ENSRNOT00000028485
|
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr10_+_65767930 | 25.03 |
ENSRNOT00000039954
|
Vtn
|
vitronectin |
| chr12_-_19114399 | 24.83 |
ENSRNOT00000073099
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr1_-_258766881 | 24.74 |
ENSRNOT00000015801
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12 |
| chr5_-_77433847 | 24.71 |
ENSRNOT00000076906
ENSRNOT00000043056 |
LOC500473
Mup4
|
similar to alpha-2u globulin PGCL2 major urinary protein 4 |
| chr11_+_74057361 | 23.79 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr10_-_71491743 | 22.86 |
ENSRNOT00000038955
|
LOC102552988
|
uncharacterized LOC102552988 |
| chr11_+_7422272 | 22.48 |
ENSRNOT00000075964
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chr5_-_173233188 | 22.46 |
ENSRNOT00000055343
|
Tmem88b
|
transmembrane protein 88B |
| chr4_+_153921900 | 22.13 |
ENSRNOT00000089482
|
Slc6a12
|
solute carrier family 6 member 12 |
| chr10_-_103340922 | 21.83 |
ENSRNOT00000004191
|
Btbd17
|
BTB domain containing 17 |
| chr4_-_62840357 | 21.82 |
ENSRNOT00000059892
|
Slc13a4
|
solute carrier family 13 member 4 |
| chr11_-_60882379 | 21.63 |
ENSRNOT00000002799
|
Cd200r1
|
CD200 receptor 1 |
| chr6_+_132242328 | 21.20 |
ENSRNOT00000081088
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr5_+_142875773 | 21.20 |
ENSRNOT00000082120
ENSRNOT00000056496 |
Epha10
|
EPH receptor A10 |
| chr9_-_19880346 | 21.17 |
ENSRNOT00000014051
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr6_+_111176798 | 21.14 |
ENSRNOT00000072215
|
Gstz1
|
glutathione S-transferase zeta 1 |
| chr5_-_140657745 | 20.95 |
ENSRNOT00000019080
|
Mfsd2a
|
major facilitator superfamily domain containing 2A |
| chr17_+_31441630 | 20.90 |
ENSRNOT00000083705
ENSRNOT00000023582 |
Tubb2b
|
tubulin, beta 2B class IIb |
| chr3_-_127500709 | 20.83 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chr5_-_4975436 | 20.83 |
ENSRNOT00000062006
|
Xkr9
|
XK related 9 |
| chr5_-_77903062 | 20.73 |
ENSRNOT00000073954
ENSRNOT00000059320 ENSRNOT00000089382 ENSRNOT00000076074 ENSRNOT00000074349 |
LOC100912565
Rn50_5_0814.4
|
major urinary protein-like alpha2u globulin (LOC298111), mRNA |
| chr4_+_109497962 | 20.69 |
ENSRNOT00000057869
|
Reg1a
|
regenerating family member 1 alpha |
| chr8_-_46537050 | 19.87 |
ENSRNOT00000011284
|
Sc5d
|
sterol-C5-desaturase |
| chr4_+_57952982 | 19.80 |
ENSRNOT00000014465
|
Cpa1
|
carboxypeptidase A1 |
| chr14_+_22251499 | 19.31 |
ENSRNOT00000087991
ENSRNOT00000002705 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr3_-_7141522 | 18.80 |
ENSRNOT00000014572
|
Cel
|
carboxyl ester lipase |
| chr2_+_150146234 | 18.70 |
ENSRNOT00000018761
|
Aadac
|
arylacetamide deacetylase |
| chr1_+_100393303 | 18.62 |
ENSRNOT00000026251
|
Syt3
|
synaptotagmin 3 |
| chr4_-_77489535 | 18.33 |
ENSRNOT00000008728
|
Pdia4
|
protein disulfide isomerase family A, member 4 |
| chr2_+_235264219 | 18.21 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr17_-_69827112 | 18.17 |
ENSRNOT00000023835
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr1_-_85220237 | 17.75 |
ENSRNOT00000026907
|
Sycn
|
syncollin |
| chr2_-_53140637 | 17.68 |
ENSRNOT00000060492
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr4_+_175729726 | 17.55 |
ENSRNOT00000013230
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr5_-_77342299 | 17.29 |
ENSRNOT00000075994
|
RGD1566134
|
similar to alpha-2u-globulin |
| chr4_+_153921115 | 17.23 |
ENSRNOT00000018821
|
Slc6a12
|
solute carrier family 6 member 12 |
| chr3_+_117416345 | 16.98 |
ENSRNOT00000056054
|
Ctxn2
|
cortexin 2 |
| chr11_-_64421248 | 16.77 |
ENSRNOT00000066997
|
Igsf11
|
immunoglobulin superfamily, member 11 |
| chr7_-_106753592 | 16.71 |
ENSRNOT00000006930
|
Kcnq3
|
potassium voltage-gated channel subfamily Q member 3 |
| chr9_+_95256627 | 16.46 |
ENSRNOT00000025291
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr3_+_134440195 | 16.43 |
ENSRNOT00000072928
|
AABR07054000.1
|
|
| chrX_+_118084890 | 16.12 |
ENSRNOT00000065293
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr5_+_124476168 | 15.96 |
ENSRNOT00000077754
|
RGD1564074
|
similar to novel protein |
| chr15_+_42659371 | 15.83 |
ENSRNOT00000091612
|
Clu
|
clusterin |
| chr14_+_2050483 | 15.56 |
ENSRNOT00000000047
|
Slc26a1
|
solute carrier family 26 member 1 |
| chr7_-_12246729 | 15.49 |
ENSRNOT00000044030
|
Reep6
|
receptor accessory protein 6 |
| chr4_+_84423653 | 15.32 |
ENSRNOT00000012655
|
Chn2
|
chimerin 2 |
| chr9_+_95285592 | 15.23 |
ENSRNOT00000063853
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr10_+_59765258 | 15.17 |
ENSRNOT00000026351
|
Shpk
|
sedoheptulokinase |
| chr16_+_3293599 | 14.81 |
ENSRNOT00000081999
ENSRNOT00000047118 ENSRNOT00000020427 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr10_-_83898527 | 14.77 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr3_+_154395187 | 14.72 |
ENSRNOT00000050810
|
Vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr5_-_58124681 | 14.67 |
ENSRNOT00000019795
|
Sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr8_+_52729003 | 14.62 |
ENSRNOT00000081738
|
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr15_+_47455690 | 14.30 |
ENSRNOT00000075276
|
LOC100910401
|
serine protease 55-like |
| chr9_-_98551410 | 14.28 |
ENSRNOT00000066346
ENSRNOT00000086678 |
Hes6
|
hes family bHLH transcription factor 6 |
| chr10_+_109708755 | 14.21 |
ENSRNOT00000083601
|
Gcgr
|
glucagon receptor |
| chr5_-_109651730 | 13.67 |
ENSRNOT00000093032
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr10_+_37215937 | 13.63 |
ENSRNOT00000006567
|
Sar1b
|
secretion associated, Ras related GTPase 1B |
| chr13_-_72063347 | 13.10 |
ENSRNOT00000090544
ENSRNOT00000003869 |
Cacna1e
|
calcium voltage-gated channel subunit alpha1 E |
| chr17_+_69588085 | 13.06 |
ENSRNOT00000064884
|
Akr1c12l1
|
aldo-keto reductase family 1, member C12-like 1 |
| chr5_-_77492013 | 13.04 |
ENSRNOT00000012293
|
LOC259245
|
alpha-2u globulin PGCL5 |
| chr1_-_211520699 | 12.67 |
ENSRNOT00000009178
|
Stk32c
|
serine/threonine kinase 32C |
| chr12_-_52124779 | 12.40 |
ENSRNOT00000088839
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr9_+_4094995 | 12.32 |
ENSRNOT00000089450
|
LOC100910057
|
sulfotransferase 1C2-like |
| chr1_-_174119815 | 12.30 |
ENSRNOT00000019368
|
Trim66
|
tripartite motif-containing 66 |
| chr15_-_14737704 | 12.15 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
| chr10_-_27366665 | 12.10 |
ENSRNOT00000004725
|
Gabra1
|
gamma-aminobutyric acid type A receptor alpha1 subunit |
| chr5_-_40237591 | 12.09 |
ENSRNOT00000011393
|
Fut9
|
fucosyltransferase 9 |
| chr1_-_220096319 | 12.02 |
ENSRNOT00000091787
ENSRNOT00000073983 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr9_+_101005082 | 11.95 |
ENSRNOT00000082949
|
Neu4
|
neuraminidase 4 |
| chr12_+_8725517 | 11.88 |
ENSRNOT00000001243
|
Slc46a3
|
solute carrier family 46, member 3 |
| chrM_+_7758 | 11.78 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr8_+_13796021 | 11.75 |
ENSRNOT00000013927
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr1_+_167937026 | 11.73 |
ENSRNOT00000020655
|
Olr57
|
olfactory receptor 57 |
| chr15_-_23969011 | 11.71 |
ENSRNOT00000014821
|
Gch1
|
GTP cyclohydrolase 1 |
| chr8_+_85259982 | 11.70 |
ENSRNOT00000010409
|
Elovl5
|
ELOVL fatty acid elongase 5 |
| chr10_-_65820538 | 11.66 |
ENSRNOT00000011808
|
Tmem97
|
transmembrane protein 97 |
| chr1_+_42170281 | 11.57 |
ENSRNOT00000092879
|
Vip
|
vasoactive intestinal peptide |
| chr2_-_188412329 | 11.49 |
ENSRNOT00000033917
ENSRNOT00000080185 |
Fdps
|
farnesyl diphosphate synthase |
| chr1_+_102900286 | 11.45 |
ENSRNOT00000017468
|
Ldha
|
lactate dehydrogenase A |
| chr5_+_78222504 | 11.36 |
ENSRNOT00000019544
|
Slc31a1
|
solute carrier family 31 member 1 |
| chr1_+_282134981 | 11.34 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr4_+_151298548 | 11.24 |
ENSRNOT00000010746
|
Cacna2d4
|
calcium voltage-gated channel auxiliary subunit alpha2delta 4 |
| chr1_-_5165859 | 11.18 |
ENSRNOT00000044325
ENSRNOT00000019319 |
Grm1
|
glutamate metabotropic receptor 1 |
| chr13_+_83681322 | 11.18 |
ENSRNOT00000004206
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr10_-_107539465 | 11.08 |
ENSRNOT00000004524
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr10_-_35833861 | 11.05 |
ENSRNOT00000049942
|
Canx
|
calnexin |
| chr3_+_114355798 | 10.98 |
ENSRNOT00000024658
ENSRNOT00000036435 |
Slc28a2
|
solute carrier family 28 member 2 |
| chr15_-_80713153 | 10.94 |
ENSRNOT00000063800
|
Klhl1
|
kelch-like family member 1 |
| chr2_-_170460754 | 10.90 |
ENSRNOT00000013009
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr13_-_36156076 | 10.89 |
ENSRNOT00000074685
|
Dbi
|
diazepam binding inhibitor, acyl-CoA binding protein |
| chr2_+_243656579 | 10.78 |
ENSRNOT00000036993
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr5_-_59553416 | 10.76 |
ENSRNOT00000090490
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr10_-_8654892 | 10.68 |
ENSRNOT00000066534
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr17_+_69468427 | 10.63 |
ENSRNOT00000058413
|
RGD1564865
|
similar to 20-alpha-hydroxysteroid dehydrogenase |
| chr13_-_50514151 | 10.56 |
ENSRNOT00000003951
|
Ren
|
renin |
| chr10_+_88997399 | 10.53 |
ENSRNOT00000027116
|
Mlx
|
MLX, MAX dimerization protein |
| chr8_-_2045817 | 10.47 |
ENSRNOT00000009542
ENSRNOT00000081171 ENSRNOT00000078765 |
Gria4
|
glutamate ionotropic receptor AMPA type subunit 4 |
| chr2_-_235177275 | 10.44 |
ENSRNOT00000093153
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr15_+_42640146 | 10.43 |
ENSRNOT00000078861
|
Clu
|
clusterin |
| chr17_+_69761118 | 10.35 |
ENSRNOT00000023739
|
Akr1c3
|
aldo-keto reductase family 1, member C3 |
| chr11_-_77593171 | 10.29 |
ENSRNOT00000002645
ENSRNOT00000043498 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr15_-_8914501 | 10.28 |
ENSRNOT00000008752
|
Thrb
|
thyroid hormone receptor beta |
| chr2_+_22909569 | 10.26 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr4_-_121969446 | 10.16 |
ENSRNOT00000085001
|
Vom1r99
|
vomeronasal 1 receptor 99 |
| chr3_-_176816114 | 10.15 |
ENSRNOT00000079262
ENSRNOT00000018697 |
Stmn3
|
stathmin 3 |
| chr9_+_95241609 | 10.12 |
ENSRNOT00000032634
ENSRNOT00000077416 |
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr5_-_166133274 | 10.11 |
ENSRNOT00000078830
|
Kif1b
|
kinesin family member 1B |
| chr16_-_81756654 | 10.08 |
ENSRNOT00000026653
|
Cul4a
|
cullin 4A |
| chr10_+_1834518 | 10.06 |
ENSRNOT00000061709
|
Gm1758
|
predicted gene 1758 |
| chr14_-_46529375 | 9.86 |
ENSRNOT00000077306
ENSRNOT00000078682 |
LOC257642
|
rRNA promoter binding protein |
| chr12_-_35979193 | 9.85 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr4_+_21462779 | 9.74 |
ENSRNOT00000089747
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr9_-_91882927 | 9.73 |
ENSRNOT00000084446
|
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr3_+_11593655 | 9.70 |
ENSRNOT00000074122
|
Pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr13_+_77485113 | 9.70 |
ENSRNOT00000080254
|
Tnr
|
tenascin R |
| chr10_+_91187593 | 9.68 |
ENSRNOT00000004163
|
Acbd4
|
acyl-CoA binding domain containing 4 |
| chr10_-_15465404 | 9.66 |
ENSRNOT00000077826
ENSRNOT00000027593 |
Decr2
|
2,4-dienoyl-CoA reductase 2 |
| chr20_+_41100071 | 9.59 |
ENSRNOT00000000658
|
Tspyl4
|
TSPY-like 4 |
| chr3_+_177310753 | 9.43 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr2_+_209840404 | 9.39 |
ENSRNOT00000050149
|
Kcna2
|
potassium voltage-gated channel subfamily A member 2 |
| chr2_-_2814690 | 9.33 |
ENSRNOT00000029265
|
Gpr150
|
G protein-coupled receptor 150 |
| chr10_-_37215899 | 9.31 |
ENSRNOT00000006356
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr1_-_264172729 | 9.25 |
ENSRNOT00000018447
|
Scd
|
stearoyl-CoA desaturase |
| chr10_-_96131880 | 9.09 |
ENSRNOT00000004578
|
Cacng5
|
calcium voltage-gated channel auxiliary subunit gamma 5 |
| chrX_+_65800059 | 9.09 |
ENSRNOT00000076428
|
Gpr165
|
G protein-coupled receptor 165 |
| chr1_-_128604188 | 9.05 |
ENSRNOT00000033858
|
Lrrc28
|
leucine rich repeat containing 28 |
| chr1_+_67025240 | 9.05 |
ENSRNOT00000051024
|
Vom1r47
|
vomeronasal 1 receptor 47 |
| chr1_-_82344345 | 9.04 |
ENSRNOT00000051892
ENSRNOT00000090629 ENSRNOT00000029487 ENSRNOT00000027933 |
Ceacam1
|
carcinoembryonic antigen related cell adhesion molecule 1 |
| chr13_-_44345735 | 9.04 |
ENSRNOT00000005006
|
Tmem163
|
transmembrane protein 163 |
| chr17_+_81455955 | 8.98 |
ENSRNOT00000044313
|
Slc39a12
|
solute carrier family 39 member 12 |
| chr10_-_107539658 | 8.97 |
ENSRNOT00000089346
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr16_-_10603850 | 8.96 |
ENSRNOT00000087737
|
Fam35a
|
family with sequence similarity 35, member A |
| chr13_-_39643361 | 8.92 |
ENSRNOT00000003527
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr5_+_135029955 | 8.91 |
ENSRNOT00000074860
|
LOC100911669
|
uncharacterized LOC100911669 |
| chr6_+_97168453 | 8.90 |
ENSRNOT00000085790
|
Syt16
|
synaptotagmin 16 |
| chr18_+_62805410 | 8.88 |
ENSRNOT00000086679
|
Gnal
|
G protein subunit alpha L |
| chr13_-_51076852 | 8.80 |
ENSRNOT00000078993
|
Adora1
|
adenosine A1 receptor |
| chrX_+_28072826 | 8.79 |
ENSRNOT00000039796
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr12_+_11814071 | 8.70 |
ENSRNOT00000039149
|
Tmem130
|
transmembrane protein 130 |
| chr6_-_115616766 | 8.66 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr2_+_228544418 | 8.64 |
ENSRNOT00000013030
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chrX_-_15707436 | 8.62 |
ENSRNOT00000085907
|
Syp
|
synaptophysin |
| chr9_+_73528681 | 8.47 |
ENSRNOT00000084340
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr4_+_22859622 | 8.46 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr1_-_213635546 | 8.45 |
ENSRNOT00000018861
|
Sirt3
|
sirtuin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cux1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 36.4 | 109.2 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 18.3 | 127.9 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 17.8 | 53.5 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 17.2 | 51.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 14.8 | 44.5 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 14.7 | 44.2 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 13.4 | 40.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 12.5 | 37.5 | GO:0033986 | response to methanol(GO:0033986) |
| 10.3 | 31.0 | GO:0042560 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 10.3 | 41.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 9.9 | 29.7 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 9.8 | 39.4 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 9.8 | 29.3 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) oxalic acid secretion(GO:0046724) |
| 9.7 | 29.2 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 8.8 | 26.3 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 8.5 | 33.8 | GO:0048252 | lauric acid metabolic process(GO:0048252) |
| 8.3 | 24.8 | GO:0009822 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 8.0 | 8.0 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 7.2 | 35.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 7.0 | 21.0 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 6.9 | 20.7 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 6.7 | 133.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 6.4 | 64.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 6.4 | 70.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 6.2 | 37.4 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 5.8 | 57.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 5.7 | 28.3 | GO:0015755 | fructose transport(GO:0015755) |
| 5.4 | 16.1 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 5.3 | 15.9 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 5.2 | 20.8 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 5.1 | 25.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 4.9 | 64.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 4.6 | 18.3 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 4.5 | 50.0 | GO:0051132 | NK T cell activation(GO:0051132) |
| 4.4 | 61.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 3.9 | 11.7 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 3.7 | 11.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 3.6 | 10.8 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 3.6 | 10.8 | GO:0046380 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 3.5 | 21.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 3.4 | 10.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 3.3 | 3.3 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 3.2 | 37.8 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 3.1 | 9.3 | GO:1903699 | tarsal gland development(GO:1903699) |
| 2.9 | 8.8 | GO:0032900 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) negative regulation of long term synaptic depression(GO:1900453) |
| 2.9 | 8.6 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 2.8 | 11.4 | GO:0098705 | copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 2.8 | 14.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 2.8 | 8.3 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 2.7 | 10.9 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 2.6 | 10.6 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 2.6 | 7.8 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 2.5 | 25.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 2.5 | 47.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 2.5 | 7.4 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 2.4 | 14.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 2.4 | 7.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 2.4 | 9.7 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 2.3 | 11.7 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 2.3 | 18.7 | GO:0070459 | prolactin secretion(GO:0070459) |
| 2.3 | 2.3 | GO:0090032 | negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 2.3 | 38.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 2.3 | 24.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 2.3 | 13.5 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 2.2 | 15.6 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 2.2 | 6.5 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 2.0 | 10.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 2.0 | 6.0 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 2.0 | 10.0 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 2.0 | 6.0 | GO:0010034 | response to acetate(GO:0010034) |
| 2.0 | 6.0 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 1.9 | 34.4 | GO:0097503 | sialylation(GO:0097503) |
| 1.9 | 5.7 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 1.9 | 11.4 | GO:0006113 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 1.9 | 5.7 | GO:0030091 | protein repair(GO:0030091) |
| 1.9 | 3.8 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 1.9 | 5.7 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 1.9 | 18.7 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 1.8 | 12.5 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 1.8 | 3.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 1.7 | 8.7 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 1.7 | 10.3 | GO:0008050 | female courtship behavior(GO:0008050) |
| 1.7 | 8.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 1.6 | 6.6 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 1.6 | 8.2 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 1.6 | 13.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 1.6 | 9.8 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 1.6 | 9.7 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.6 | 8.0 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.6 | 49.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 1.6 | 4.7 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 1.6 | 9.4 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 1.6 | 39.0 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 1.6 | 9.3 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 1.5 | 3.0 | GO:0097187 | dentinogenesis(GO:0097187) |
| 1.5 | 8.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 1.4 | 4.3 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 1.4 | 55.5 | GO:0006956 | complement activation(GO:0006956) |
| 1.4 | 6.9 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 1.4 | 5.5 | GO:0014028 | notochord formation(GO:0014028) |
| 1.4 | 6.8 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 1.3 | 6.7 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 1.3 | 3.9 | GO:0001966 | thigmotaxis(GO:0001966) |
| 1.3 | 14.2 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 1.3 | 3.8 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 1.3 | 8.9 | GO:0009405 | pathogenesis(GO:0009405) |
| 1.3 | 3.8 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 1.3 | 3.8 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 1.2 | 2.5 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 1.2 | 6.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.2 | 4.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.2 | 29.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 1.2 | 15.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.2 | 4.7 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 1.2 | 8.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 1.2 | 8.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.2 | 2.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 1.1 | 21.8 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 1.1 | 7.7 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 1.1 | 29.0 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 1.1 | 3.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 1.1 | 9.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 1.0 | 7.3 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 1.0 | 9.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 1.0 | 8.4 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 1.0 | 3.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 1.0 | 5.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 1.0 | 5.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.0 | 12.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.0 | 3.9 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 1.0 | 2.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 1.0 | 44.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 1.0 | 9.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.9 | 3.8 | GO:0071283 | cellular response to iron(III) ion(GO:0071283) |
| 0.9 | 2.8 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.9 | 4.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.9 | 3.7 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.9 | 2.8 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.9 | 5.5 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.9 | 2.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.9 | 2.7 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.9 | 5.4 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.9 | 3.6 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.9 | 9.8 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.9 | 4.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.9 | 3.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.9 | 25.4 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.9 | 8.7 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.9 | 12.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.9 | 1.7 | GO:1902741 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.8 | 8.5 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.8 | 0.8 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.8 | 9.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.8 | 2.4 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.8 | 6.3 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.8 | 45.8 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.8 | 2.4 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.8 | 6.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.8 | 13.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.8 | 16.7 | GO:0061548 | ganglion development(GO:0061548) |
| 0.7 | 6.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.7 | 3.7 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.7 | 6.6 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.7 | 7.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.7 | 5.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.7 | 3.5 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.7 | 2.1 | GO:2000853 | negative regulation of corticosterone secretion(GO:2000853) |
| 0.7 | 8.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.6 | 3.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.6 | 17.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.6 | 1.8 | GO:0060125 | pancreatic amylase secretion(GO:0036395) negative regulation of growth hormone secretion(GO:0060125) regulation of pancreatic amylase secretion(GO:1902276) |
| 0.6 | 7.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.6 | 5.4 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.6 | 1.8 | GO:0090648 | response to environmental enrichment(GO:0090648) |
| 0.6 | 10.7 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.6 | 5.3 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.6 | 2.9 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.6 | 6.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.6 | 2.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.6 | 17.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.6 | 3.3 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.6 | 2.8 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.5 | 10.4 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.5 | 9.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.5 | 4.8 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.5 | 1.6 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.5 | 1.6 | GO:2000464 | positive regulation of astrocyte chemotaxis(GO:2000464) |
| 0.5 | 8.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.5 | 4.5 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.5 | 2.0 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.5 | 1.5 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.5 | 2.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.5 | 1.5 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.5 | 3.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.5 | 12.0 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.5 | 6.7 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.5 | 1.4 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.5 | 3.7 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.5 | 3.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.4 | 2.2 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.4 | 0.9 | GO:1904954 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.4 | 2.6 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.4 | 11.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.4 | 6.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.4 | 2.9 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.4 | 35.2 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.4 | 16.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.4 | 4.8 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.4 | 2.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.4 | 1.9 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) spermine catabolic process(GO:0046208) |
| 0.4 | 5.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.4 | 1.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.4 | 17.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.4 | 3.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.4 | 6.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.4 | 2.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.4 | 1.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.4 | 8.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.3 | 4.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.3 | 4.5 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 4.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 65.7 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.3 | 9.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.3 | 5.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.3 | 13.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.3 | 0.3 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.3 | 28.5 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.3 | 0.9 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.3 | 9.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.3 | 3.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 11.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.3 | 1.2 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.3 | 1.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 2.6 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.3 | 1.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.3 | 5.3 | GO:0050951 | sensory perception of temperature stimulus(GO:0050951) |
| 0.3 | 3.9 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 1.9 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.3 | 1.6 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.3 | 1.3 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.3 | 5.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.3 | 4.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 1.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.3 | 1.5 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.3 | 5.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.3 | 3.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.2 | 1.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 3.1 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.2 | 3.6 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 3.5 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 3.0 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 4.1 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.2 | 2.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 1.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 2.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 5.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 3.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 6.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 254.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 0.4 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.2 | 0.9 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 0.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 9.4 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.2 | 12.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 1.6 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.2 | 6.2 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 4.8 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.2 | 9.0 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.2 | 5.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 2.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.2 | 1.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 4.7 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 0.8 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.2 | 4.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 3.9 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.1 | 2.2 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.6 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 1.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.4 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.1 | 0.6 | GO:0090181 | regulation of cholesterol metabolic process(GO:0090181) |
| 0.1 | 1.6 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 1.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.1 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 0.6 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 2.4 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 1.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.8 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 7.5 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.1 | 0.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 4.8 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 1.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.4 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 1.3 | GO:0015671 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.1 | 1.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.7 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.1 | 0.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.7 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 3.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 6.0 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 0.3 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.1 | 2.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.5 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.1 | 1.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 0.2 | GO:2000292 | regulation of eating behavior(GO:1903998) negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 0.9 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 0.9 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 1.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 1.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 2.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 1.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.3 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.4 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 1.4 | GO:0001942 | hair follicle development(GO:0001942) |
| 0.0 | 0.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.0 | 0.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.0 | 98.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 11.7 | 70.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 5.2 | 15.5 | GO:0044317 | rod spherule(GO:0044317) |
| 4.8 | 52.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 2.9 | 8.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 2.8 | 17.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 2.8 | 11.2 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 2.8 | 8.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 2.4 | 7.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 2.4 | 47.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 2.3 | 6.9 | GO:0060187 | cell pole(GO:0060187) |
| 2.2 | 6.5 | GO:0071942 | XPC complex(GO:0071942) |
| 2.1 | 44.5 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 2.1 | 27.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.9 | 9.7 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.7 | 18.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.6 | 6.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.5 | 7.7 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.4 | 29.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.2 | 4.8 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 1.2 | 7.0 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 1.1 | 11.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 1.1 | 10.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.1 | 11.0 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 1.1 | 10.7 | GO:0030891 | VCB complex(GO:0030891) |
| 1.1 | 18.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 1.0 | 5.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 1.0 | 124.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.9 | 26.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.9 | 3.5 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.8 | 2.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.8 | 4.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.8 | 40.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.7 | 113.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.7 | 19.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.7 | 2.8 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.7 | 8.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.7 | 11.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.7 | 15.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.6 | 1.2 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.6 | 28.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.6 | 12.1 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.6 | 36.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.6 | 9.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.5 | 9.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.5 | 6.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.5 | 20.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.5 | 7.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.5 | 3.8 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.5 | 9.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.5 | 1.5 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.5 | 7.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.5 | 2.5 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.5 | 361.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.5 | 9.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.4 | 1.8 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.4 | 2.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.4 | 12.6 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.4 | 3.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.4 | 1.6 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.4 | 9.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.4 | 3.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.3 | 27.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.3 | 1.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 2.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.3 | 4.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 18.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 6.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 28.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.3 | 1.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.3 | 2.7 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.3 | 34.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.3 | 1.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 10.8 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 1.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.3 | 18.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 80.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.3 | 3.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.3 | 2.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 6.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.2 | 2.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 2.8 | GO:0030666 | endocytic vesicle membrane(GO:0030666) phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 8.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 0.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.2 | 9.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 6.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 2.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 3.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 292.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 0.6 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 1.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 6.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 1.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 2.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 12.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 7.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 3.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 9.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.5 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 2.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 10.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 0.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 5.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 5.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 3.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 58.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 72.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 7.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 13.4 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 184.5 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.0 | 79.8 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 15.6 | 62.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 12.5 | 37.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 12.4 | 37.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 11.7 | 11.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 11.4 | 57.2 | GO:0019862 | IgA binding(GO:0019862) |
| 10.8 | 53.8 | GO:0005534 | galactose binding(GO:0005534) |
| 10.3 | 51.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 10.2 | 335.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) |
| 9.8 | 49.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 9.3 | 56.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 8.4 | 42.0 | GO:0005550 | pheromone binding(GO:0005550) |
| 8.3 | 33.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 7.5 | 22.5 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 7.1 | 28.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 7.1 | 21.2 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 6.6 | 26.6 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 6.6 | 39.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 6.3 | 18.8 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 6.2 | 192.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 5.9 | 29.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 5.9 | 29.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 5.4 | 16.1 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 5.2 | 20.8 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) |
| 5.1 | 15.3 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 5.1 | 25.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 4.4 | 31.0 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 4.2 | 29.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 3.6 | 10.9 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 3.6 | 25.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 3.6 | 10.8 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 3.6 | 17.8 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 3.6 | 14.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 3.5 | 10.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 3.5 | 52.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 3.4 | 10.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 3.2 | 9.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 3.2 | 16.1 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 3.2 | 35.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 3.2 | 25.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 3.1 | 24.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 3.0 | 27.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 3.0 | 14.9 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 2.9 | 11.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 2.8 | 67.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 2.7 | 11.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 2.7 | 35.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 2.6 | 13.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 2.6 | 15.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 2.4 | 14.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 2.4 | 29.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 2.4 | 12.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 2.4 | 40.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 2.3 | 11.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 2.2 | 11.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 2.2 | 39.5 | GO:0001848 | complement binding(GO:0001848) |
| 2.1 | 8.6 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 2.0 | 12.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 2.0 | 10.0 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 2.0 | 6.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.9 | 5.7 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.9 | 15.0 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 1.8 | 7.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 1.8 | 9.0 | GO:0046790 | virion binding(GO:0046790) |
| 1.8 | 10.8 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.7 | 21.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 1.7 | 45.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.7 | 10.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 1.6 | 8.2 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 1.6 | 26.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 1.6 | 6.5 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 1.6 | 8.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.6 | 4.7 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 1.6 | 4.7 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 1.5 | 6.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 1.5 | 21.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 1.5 | 6.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.5 | 8.9 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 1.5 | 4.4 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 1.4 | 5.7 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 1.4 | 5.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 1.4 | 11.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 1.4 | 4.3 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 1.4 | 11.2 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 1.4 | 6.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 1.4 | 6.8 | GO:0043559 | insulin binding(GO:0043559) |
| 1.4 | 29.9 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 1.3 | 6.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.2 | 20.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 1.2 | 4.8 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 1.2 | 3.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.2 | 4.8 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 1.1 | 16.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.1 | 4.4 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 1.0 | 3.9 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 1.0 | 3.8 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.9 | 10.3 | GO:0070324 | thyroid hormone receptor activity(GO:0004887) thyroid hormone binding(GO:0070324) |
| 0.9 | 3.7 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.9 | 2.7 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.9 | 3.6 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.9 | 8.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.9 | 14.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.9 | 8.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.9 | 18.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.8 | 6.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.8 | 197.2 | GO:0017171 | serine hydrolase activity(GO:0017171) |
| 0.8 | 4.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.8 | 27.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.8 | 3.3 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.8 | 7.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.8 | 8.0 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.8 | 4.7 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.8 | 7.0 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.8 | 3.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.8 | 8.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.8 | 3.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.7 | 11.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.7 | 14.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.7 | 3.5 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.7 | 8.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.7 | 31.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.7 | 9.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.6 | 1.9 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.6 | 2.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.6 | 5.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.6 | 54.9 | GO:0005496 | steroid binding(GO:0005496) |
| 0.6 | 21.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.6 | 2.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.6 | 9.8 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.6 | 4.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.6 | 5.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.5 | 14.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.5 | 7.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.5 | 1.6 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.5 | 3.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.5 | 1.5 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.5 | 2.5 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.5 | 5.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.5 | 2.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.5 | 1.4 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.5 | 9.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.5 | 3.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.5 | 5.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.4 | 1.8 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.4 | 3.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.4 | 20.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.4 | 9.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.4 | 2.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.4 | 8.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.4 | 1.2 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.4 | 3.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.4 | 8.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.4 | 8.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 3.5 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 1.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.3 | 3.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 2.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.3 | 13.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.3 | 1.6 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.3 | 2.2 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 6.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 1.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.3 | 1.1 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 3.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 1.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 2.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.3 | 1.0 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.2 | 14.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 4.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 4.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 4.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 3.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 2.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.2 | 1.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 0.8 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 4.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 3.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.2 | 4.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 8.2 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 255.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 5.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 0.9 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.2 | 16.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 21.3 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.2 | 8.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 0.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.2 | 16.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 4.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 5.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 16.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.2 | 3.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 3.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 6.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 18.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 3.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 3.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 4.8 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 3.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 2.6 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 2.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 4.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 7.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 0.4 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 0.9 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.3 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 2.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.3 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.6 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 70.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 1.9 | 20.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 1.4 | 343.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.8 | 15.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.7 | 37.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.6 | 10.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.6 | 27.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 4.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.5 | 5.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.4 | 7.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 8.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 7.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.3 | 49.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 14.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 10.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.3 | 14.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 13.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 10.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 9.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 15.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.2 | 5.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 12.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 4.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 10.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 2.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 1.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 7.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 3.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 148.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 5.1 | 97.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 4.6 | 97.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 4.6 | 160.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 4.2 | 37.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 2.5 | 66.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 2.5 | 32.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 2.1 | 49.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 1.8 | 17.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 1.6 | 53.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 1.6 | 27.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.5 | 21.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.5 | 8.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 1.5 | 11.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.5 | 26.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 1.4 | 24.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 1.4 | 6.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 1.3 | 8.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 1.3 | 22.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 1.2 | 35.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 1.0 | 11.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 1.0 | 20.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.9 | 151.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.9 | 12.2 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.8 | 9.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.8 | 71.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.8 | 32.9 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.7 | 57.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.7 | 12.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.7 | 10.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.7 | 12.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.7 | 6.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.6 | 14.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.6 | 11.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.5 | 10.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.5 | 8.8 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.5 | 26.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.5 | 6.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 14.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.5 | 25.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 8.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 8.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.4 | 6.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.4 | 6.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 1.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.4 | 26.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 3.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 6.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 6.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.3 | 12.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 5.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.3 | 3.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 6.6 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 4.0 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 2.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 6.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 5.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 6.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 3.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 4.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 5.5 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.2 | 10.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 3.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 5.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 4.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.0 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 4.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 2.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |