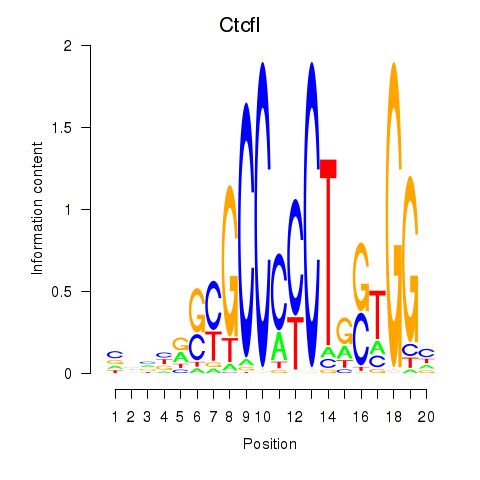
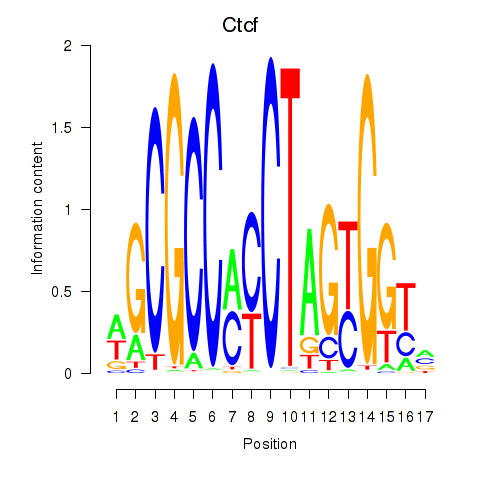
Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Ctcfl_Ctcf
Z-value: 1.15
Transcription factors associated with Ctcfl_Ctcf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ctcfl
|
ENSRNOG00000028661 | CCCTC-binding factor like |
|
Ctcf
|
ENSRNOG00000017674 | CCCTC-binding factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ctcf | rn6_v1_chr19_+_37600148_37600148 | 0.16 | 4.2e-03 | Click! |
| Ctcfl | rn6_v1_chr3_-_171166454_171166454 | -0.13 | 2.2e-02 | Click! |
Activity profile of Ctcfl_Ctcf motif
Sorted Z-values of Ctcfl_Ctcf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_41482728 | 73.62 |
ENSRNOT00000022943
|
Calb2
|
calbindin 2 |
| chr3_-_11382004 | 59.90 |
ENSRNOT00000047921
ENSRNOT00000064039 |
Dnm1
|
dynamin 1 |
| chr3_-_52510507 | 57.80 |
ENSRNOT00000091259
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr14_+_83752393 | 57.32 |
ENSRNOT00000081123
|
Selenom
|
selenoprotein M |
| chr3_+_51687809 | 55.89 |
ENSRNOT00000087242
|
Scn2a
|
sodium voltage-gated channel alpha subunit 2 |
| chr9_-_32019205 | 44.96 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr3_-_51643140 | 44.85 |
ENSRNOT00000006646
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr1_-_89483988 | 44.55 |
ENSRNOT00000028603
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr11_+_36851038 | 43.86 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr18_+_30004565 | 43.57 |
ENSRNOT00000027393
|
Pcdha4
|
protocadherin alpha 4 |
| chr6_+_137997335 | 42.86 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr18_+_29951094 | 42.14 |
ENSRNOT00000027402
|
LOC102553180
|
protocadherin alpha-1-like |
| chr18_+_30509393 | 36.58 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr12_-_41448668 | 36.02 |
ENSRNOT00000001856
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr18_+_30023828 | 35.52 |
ENSRNOT00000079008
|
Pcdha4
|
protocadherin alpha 4 |
| chr18_+_30474947 | 34.96 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chr8_-_117237229 | 33.96 |
ENSRNOT00000071381
|
Klhdc8b
|
kelch domain containing 8B |
| chr18_+_29972808 | 33.87 |
ENSRNOT00000074051
|
Pcdha4
|
protocadherin alpha 4 |
| chr1_+_241594565 | 33.40 |
ENSRNOT00000020123
|
Apba1
|
amyloid beta precursor protein binding family A member 1 |
| chr10_-_90393317 | 32.22 |
ENSRNOT00000028563
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr1_+_28454966 | 32.06 |
ENSRNOT00000078841
ENSRNOT00000030327 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr1_+_220335254 | 31.89 |
ENSRNOT00000072261
|
Rin1
|
Ras and Rab interactor 1 |
| chr3_-_10602672 | 30.55 |
ENSRNOT00000011648
|
Ncs1
|
neuronal calcium sensor 1 |
| chr10_-_83332851 | 30.53 |
ENSRNOT00000007133
|
Nxph3
|
neurexophilin 3 |
| chr1_+_89491654 | 30.52 |
ENSRNOT00000028632
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr4_-_119131202 | 30.17 |
ENSRNOT00000011675
|
Antxr1
|
anthrax toxin receptor 1 |
| chr18_+_29966245 | 29.63 |
ENSRNOT00000074028
|
Pcdha4
|
protocadherin alpha 4 |
| chr3_-_39596718 | 29.28 |
ENSRNOT00000006784
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr2_-_44907030 | 29.02 |
ENSRNOT00000013979
|
Gpx8
|
glutathione peroxidase 8 |
| chr10_+_92288910 | 28.12 |
ENSRNOT00000006947
ENSRNOT00000045127 |
Mapt
|
microtubule-associated protein tau |
| chr12_+_14092541 | 27.84 |
ENSRNOT00000033998
|
Radil
|
Rap associating with DIL domain |
| chr1_+_220428481 | 27.30 |
ENSRNOT00000027335
|
LOC108348044
|
ras and Rab interactor 1 |
| chr7_-_140483693 | 26.48 |
ENSRNOT00000089060
|
Ddn
|
dendrin |
| chr12_-_30566032 | 25.16 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr1_+_221792221 | 24.85 |
ENSRNOT00000054828
|
Nrxn2
|
neurexin 2 |
| chr11_+_86516390 | 24.63 |
ENSRNOT00000041168
|
Gp1bb
|
glycoprotein Ib platelet beta subunit |
| chr18_+_29993361 | 24.34 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chr1_-_226924244 | 23.59 |
ENSRNOT00000028971
|
Tmem132a
|
transmembrane protein 132A |
| chr7_+_123510804 | 23.56 |
ENSRNOT00000010491
|
Sept3
|
septin 3 |
| chr10_-_97582188 | 23.16 |
ENSRNOT00000005076
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr10_+_86303727 | 21.64 |
ENSRNOT00000037752
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr1_-_226353611 | 21.51 |
ENSRNOT00000037624
|
Dagla
|
diacylglycerol lipase, alpha |
| chr16_+_21282467 | 21.43 |
ENSRNOT00000065345
|
Yjefn3
|
YjeF N-terminal domain containing 3 |
| chr10_+_108395860 | 20.90 |
ENSRNOT00000075796
|
Gaa
|
glucosidase, alpha, acid |
| chr8_-_115981910 | 20.86 |
ENSRNOT00000019867
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr10_+_92289107 | 20.83 |
ENSRNOT00000050070
|
Mapt
|
microtubule-associated protein tau |
| chr1_-_73753128 | 20.66 |
ENSRNOT00000068459
|
Ttyh1
|
tweety family member 1 |
| chr3_+_154395187 | 20.58 |
ENSRNOT00000050810
|
Vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr10_+_91187593 | 20.02 |
ENSRNOT00000004163
|
Acbd4
|
acyl-CoA binding domain containing 4 |
| chr1_+_40389638 | 19.89 |
ENSRNOT00000021471
|
Plekhg1
|
pleckstrin homology and RhoGEF domain containing G1 |
| chr18_+_30515962 | 19.70 |
ENSRNOT00000027172
|
LOC108348233
|
protocadherin beta-6-like |
| chr11_+_73738433 | 19.57 |
ENSRNOT00000002353
|
Tmem44
|
transmembrane protein 44 |
| chr4_+_25825567 | 19.16 |
ENSRNOT00000009417
|
Cdk14
|
cyclin-dependent kinase 14 |
| chr10_-_88266210 | 18.84 |
ENSRNOT00000090702
ENSRNOT00000020603 |
Hap1
|
huntingtin-associated protein 1 |
| chr12_-_23925741 | 18.83 |
ENSRNOT00000001957
|
Srrm3
|
serine/arginine repetitive matrix 3 |
| chr7_-_117880289 | 18.70 |
ENSRNOT00000052227
|
Arhgap39
|
Rho GTPase activating protein 39 |
| chr18_+_30550877 | 18.69 |
ENSRNOT00000027164
|
LOC108348201
|
protocadherin beta-7-like |
| chr18_+_30527705 | 18.57 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr12_+_24651314 | 18.07 |
ENSRNOT00000077016
ENSRNOT00000071569 |
Vps37d
|
VPS37D, ESCRT-I subunit |
| chr2_+_174013288 | 17.70 |
ENSRNOT00000013904
|
Serpini1
|
serpin family I member 1 |
| chr10_-_31041626 | 17.67 |
ENSRNOT00000007267
|
Lsm11
|
LSM11, U7 small nuclear RNA associated |
| chr16_+_81616604 | 17.66 |
ENSRNOT00000026392
ENSRNOT00000057740 |
Adprhl1
Grtp1
|
ADP-ribosylhydrolase like 1 growth hormone regulated TBC protein 1 |
| chr18_+_30036887 | 17.64 |
ENSRNOT00000077824
|
Pcdha4
|
protocadherin alpha 4 |
| chrX_-_131343853 | 17.47 |
ENSRNOT00000038653
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr8_+_119566509 | 17.08 |
ENSRNOT00000028633
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr9_+_10941613 | 16.57 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr1_-_146226462 | 16.52 |
ENSRNOT00000036343
|
Cemip
|
cell migration-inducing hyaluronan binding protein |
| chr17_-_9695292 | 16.46 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr3_-_80873887 | 16.41 |
ENSRNOT00000024280
|
Dgkz
|
diacylglycerol kinase zeta |
| chr5_+_79055521 | 16.31 |
ENSRNOT00000010333
|
Col27a1
|
collagen type XXVII alpha 1 chain |
| chr1_-_226292480 | 15.65 |
ENSRNOT00000039133
|
Myrf
|
myelin regulatory factor |
| chr19_+_52086325 | 15.64 |
ENSRNOT00000020341
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr8_+_128972311 | 15.24 |
ENSRNOT00000025460
|
Myrip
|
myosin VIIA and Rab interacting protein |
| chr3_-_57957346 | 15.19 |
ENSRNOT00000036728
|
Slc25a12
|
solute carrier family 25 member 12 |
| chr4_+_34962339 | 15.15 |
ENSRNOT00000086248
|
AABR07059778.1
|
|
| chr19_-_56311991 | 14.80 |
ENSRNOT00000014090
|
Dbndd1
|
dysbindin domain containing 1 |
| chr13_+_82072497 | 14.74 |
ENSRNOT00000063810
ENSRNOT00000085135 |
Kifap3
|
kinesin-associated protein 3 |
| chr18_-_37776453 | 14.44 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chrX_-_105568343 | 14.31 |
ENSRNOT00000029807
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr14_+_79538911 | 14.30 |
ENSRNOT00000009960
|
Sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr7_+_140788987 | 14.23 |
ENSRNOT00000086611
|
Kcnh3
|
potassium voltage-gated channel subfamily H member 3 |
| chr10_+_35392762 | 14.23 |
ENSRNOT00000059277
|
Rasgef1c
|
RasGEF domain family, member 1C |
| chr8_-_110813000 | 14.08 |
ENSRNOT00000010634
|
Ephb1
|
Eph receptor B1 |
| chr5_-_166133274 | 14.06 |
ENSRNOT00000078830
|
Kif1b
|
kinesin family member 1B |
| chr5_-_166133491 | 13.99 |
ENSRNOT00000087739
ENSRNOT00000089099 |
Kif1b
|
kinesin family member 1B |
| chr9_+_117583610 | 13.81 |
ENSRNOT00000088647
ENSRNOT00000049426 |
Epb41l3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr19_-_26053762 | 13.73 |
ENSRNOT00000004646
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr12_+_47551935 | 13.58 |
ENSRNOT00000056932
|
RGD1560398
|
RGD1560398 |
| chr2_+_226900619 | 13.51 |
ENSRNOT00000019638
|
Pde5a
|
phosphodiesterase 5A |
| chr1_+_78818404 | 13.44 |
ENSRNOT00000090417
|
Gng8
|
G protein subunit gamma 8 |
| chr10_-_68517564 | 13.30 |
ENSRNOT00000086961
|
Asic2
|
acid sensing ion channel subunit 2 |
| chr8_-_47094352 | 13.20 |
ENSRNOT00000048347
|
Grik4
|
glutamate ionotropic receptor kainate type subunit 4 |
| chr3_-_148493225 | 13.04 |
ENSRNOT00000012141
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr5_-_135025084 | 13.01 |
ENSRNOT00000018766
|
Tspan1
|
tetraspanin 1 |
| chr7_-_70630338 | 12.96 |
ENSRNOT00000009803
|
Gli1
|
GLI family zinc finger 1 |
| chr2_-_80667481 | 12.96 |
ENSRNOT00000016784
|
Trio
|
trio Rho guanine nucleotide exchange factor |
| chr2_+_207923775 | 12.89 |
ENSRNOT00000019997
ENSRNOT00000051835 |
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr13_-_74923402 | 12.81 |
ENSRNOT00000033324
|
Rasal2
|
RAS protein activator like 2 |
| chr3_-_9326993 | 12.67 |
ENSRNOT00000090137
|
Lamc3
|
laminin subunit gamma 3 |
| chr2_-_188561267 | 12.56 |
ENSRNOT00000089781
ENSRNOT00000092093 |
Trim46
|
tripartite motif-containing 46 |
| chr9_-_19749145 | 12.40 |
ENSRNOT00000013956
|
Rcan2
|
regulator of calcineurin 2 |
| chr12_+_18074033 | 12.39 |
ENSRNOT00000001727
|
LOC103689975
|
integrator complex subunit 1-like |
| chrX_+_6273733 | 12.28 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr13_+_110920830 | 12.08 |
ENSRNOT00000077014
ENSRNOT00000076362 |
Kcnh1
|
potassium voltage-gated channel subfamily H member 1 |
| chr10_+_105861743 | 12.05 |
ENSRNOT00000064410
|
Mgat5b
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr6_-_91250138 | 11.90 |
ENSRNOT00000052408
|
LOC100911256
|
ninein-like |
| chr3_-_2803574 | 11.77 |
ENSRNOT00000040995
|
RGD1560470
|
similar to Gene model 996 |
| chr16_-_24788740 | 11.66 |
ENSRNOT00000018952
|
Npy1r
|
neuropeptide Y receptor Y1 |
| chr3_+_146546387 | 11.64 |
ENSRNOT00000009946
|
Entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 |
| chr11_+_36497509 | 11.55 |
ENSRNOT00000002222
|
Wrb
|
tryptophan rich basic protein |
| chr10_+_40543288 | 11.55 |
ENSRNOT00000016755
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr4_-_171176581 | 11.49 |
ENSRNOT00000030850
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr1_-_219532609 | 11.26 |
ENSRNOT00000025641
|
Ankrd13d
|
ankyrin repeat domain 13D |
| chr4_+_140837745 | 11.21 |
ENSRNOT00000080663
|
Arl8b
|
ADP-ribosylation factor like GTPase 8B |
| chr8_-_65587427 | 11.10 |
ENSRNOT00000016491
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr10_+_764421 | 11.07 |
ENSRNOT00000084608
ENSRNOT00000087567 |
Myh11
|
myosin heavy chain 11 |
| chr7_-_12793711 | 11.01 |
ENSRNOT00000013762
|
Palm
|
paralemmin |
| chr8_-_72836159 | 11.00 |
ENSRNOT00000024617
|
Tpm1
|
tropomyosin 1, alpha |
| chr1_+_127802978 | 10.96 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr10_+_69737328 | 10.83 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr3_+_123754057 | 10.83 |
ENSRNOT00000034201
ENSRNOT00000084671 |
Ap5s1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr1_-_254671596 | 10.75 |
ENSRNOT00000025450
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr2_+_195719543 | 10.74 |
ENSRNOT00000028324
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr3_+_146980923 | 10.67 |
ENSRNOT00000011654
|
Nsfl1c
|
NSFL1 cofactor |
| chr13_+_67611708 | 10.57 |
ENSRNOT00000063833
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr6_-_128989812 | 10.47 |
ENSRNOT00000085943
|
LOC100909439
|
ankyrin repeat and SOCS box protein 2-like |
| chr18_+_30487264 | 10.46 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr7_+_34001506 | 10.41 |
ENSRNOT00000005762
|
Cdk17
|
cyclin-dependent kinase 17 |
| chr13_-_51992693 | 10.38 |
ENSRNOT00000008282
|
Gpr37l1
|
G protein-coupled receptor 37-like 1 |
| chr16_+_81204766 | 10.37 |
ENSRNOT00000042465
|
Tmem255b
|
transmembrane protein 255B |
| chr12_+_49328977 | 10.35 |
ENSRNOT00000000898
|
Sgsm1
|
small G protein signaling modulator 1 |
| chr2_-_198834038 | 10.31 |
ENSRNOT00000031484
|
Nudt17
|
nudix hydrolase 17 |
| chr7_+_142397371 | 9.97 |
ENSRNOT00000040890
ENSRNOT00000065379 |
Slc4a8
|
solute carrier family 4 member 8 |
| chr1_+_47605415 | 9.94 |
ENSRNOT00000034842
|
Fndc1
|
fibronectin type III domain containing 1 |
| chr4_+_77519028 | 9.81 |
ENSRNOT00000008743
|
Zfp398
|
zinc finger protein 398 |
| chr17_+_31493107 | 9.75 |
ENSRNOT00000023611
ENSRNOT00000086264 |
Tubb2a
|
tubulin, beta 2A class IIa |
| chr17_+_77601877 | 9.67 |
ENSRNOT00000091554
ENSRNOT00000024872 |
Prpf18
|
pre-mRNA processing factor 18 |
| chr6_-_137733026 | 9.57 |
ENSRNOT00000019213
|
Jag2
|
jagged 2 |
| chr4_-_82295470 | 9.50 |
ENSRNOT00000091073
|
Hoxa10
|
homeobox A10 |
| chr1_+_261415191 | 9.47 |
ENSRNOT00000083287
ENSRNOT00000040740 |
Zfyve27
|
zinc finger FYVE-type containing 27 |
| chr1_+_47605262 | 9.46 |
ENSRNOT00000089458
|
Fndc1
|
fibronectin type III domain containing 1 |
| chr13_-_51076165 | 9.44 |
ENSRNOT00000004602
|
Adora1
|
adenosine A1 receptor |
| chr6_-_45629168 | 9.41 |
ENSRNOT00000009877
|
Rnf144a
|
ring finger protein 144A |
| chr17_+_11953552 | 9.38 |
ENSRNOT00000090782
|
Ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr14_+_77712240 | 9.37 |
ENSRNOT00000009101
|
Msx1
|
msh homeobox 1 |
| chr7_+_141702038 | 9.32 |
ENSRNOT00000086577
|
Dip2b
|
disco-interacting protein 2 homolog B |
| chr19_-_37245217 | 9.30 |
ENSRNOT00000020927
|
MGC116202
|
hypothetical protein LOC688735 |
| chr11_+_84396033 | 9.22 |
ENSRNOT00000002316
|
Abcc5
|
ATP binding cassette subfamily C member 5 |
| chr12_+_22026075 | 9.10 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr1_-_254671778 | 9.03 |
ENSRNOT00000025493
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr16_-_54450426 | 8.95 |
ENSRNOT00000014623
|
Pdgfrl
|
platelet-derived growth factor receptor-like |
| chr4_-_82209933 | 8.94 |
ENSRNOT00000091106
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr3_+_2648885 | 8.94 |
ENSRNOT00000020339
|
Abca2
|
ATP binding cassette subfamily A member 2 |
| chr1_-_85517360 | 8.86 |
ENSRNOT00000026114
|
Eid2
|
EP300 interacting inhibitor of differentiation 2 |
| chr2_+_251817694 | 8.85 |
ENSRNOT00000019964
|
RGD1560065
|
similar to RIKEN cDNA 2410004B18 |
| chr8_-_62987182 | 8.82 |
ENSRNOT00000070885
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr9_-_15306465 | 8.71 |
ENSRNOT00000019404
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr10_-_57309298 | 8.69 |
ENSRNOT00000056622
|
Camta2
|
calmodulin binding transcription activator 2 |
| chr3_-_110140756 | 8.66 |
ENSRNOT00000007882
|
Gpr176
|
G protein-coupled receptor 176 |
| chr7_-_125407806 | 8.66 |
ENSRNOT00000084666
|
RGD1566029
|
similar to mKIAA1644 protein |
| chr1_+_101427195 | 8.64 |
ENSRNOT00000028271
|
Gys1
|
glycogen synthase 1 |
| chr13_+_110257571 | 8.62 |
ENSRNOT00000005715
|
Ints7
|
integrator complex subunit 7 |
| chr18_+_30913842 | 8.60 |
ENSRNOT00000026947
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3 |
| chr5_+_104941066 | 8.59 |
ENSRNOT00000009225
|
Rraga
|
Ras-related GTP binding A |
| chr14_-_84170301 | 8.57 |
ENSRNOT00000080413
|
Slc35e4
|
solute carrier family 35, member E4 |
| chr3_+_2512492 | 8.54 |
ENSRNOT00000074026
|
AABR07051241.1
|
|
| chr5_+_70441123 | 8.42 |
ENSRNOT00000087517
|
Fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chrX_-_35431164 | 8.40 |
ENSRNOT00000004968
|
Scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr8_-_49045154 | 8.38 |
ENSRNOT00000088034
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr10_+_48240127 | 8.32 |
ENSRNOT00000080682
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr11_-_88343627 | 8.28 |
ENSRNOT00000002530
|
Ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr7_+_12022285 | 8.22 |
ENSRNOT00000024080
|
Rexo1
|
RNA exonuclease 1 homolog |
| chr1_+_218466289 | 8.19 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr14_-_18076907 | 8.15 |
ENSRNOT00000003472
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr1_-_100671074 | 7.93 |
ENSRNOT00000027132
|
Myh14
|
myosin heavy chain 14 |
| chr18_+_30375798 | 7.92 |
ENSRNOT00000060497
|
Pcdhb2
|
protocadherin beta 2 |
| chr6_+_136720266 | 7.85 |
ENSRNOT00000018278
|
Kif26a
|
kinesin family member 26A |
| chr7_+_47928060 | 7.84 |
ENSRNOT00000005740
|
Ccdc59
|
coiled-coil domain containing 59 |
| chr10_+_48240330 | 7.75 |
ENSRNOT00000057798
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr7_-_83701447 | 7.70 |
ENSRNOT00000088893
|
AABR07057675.1
|
|
| chr4_-_155561324 | 7.67 |
ENSRNOT00000085447
|
Slc2a3
|
solute carrier family 2 member 3 |
| chr1_-_213981317 | 7.67 |
ENSRNOT00000086238
ENSRNOT00000054869 |
Slc25a22
|
solute carrier family 25 member 22 |
| chr9_-_93735636 | 7.63 |
ENSRNOT00000025582
|
Nppc
|
natriuretic peptide C |
| chr2_-_187133993 | 7.62 |
ENSRNOT00000019502
|
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr1_+_40529045 | 7.47 |
ENSRNOT00000026564
|
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr11_-_65350442 | 7.46 |
ENSRNOT00000003773
|
Gpr156
|
G protein-coupled receptor 156 |
| chr17_-_79676499 | 7.46 |
ENSRNOT00000022711
|
Itga8
|
integrin subunit alpha 8 |
| chr9_+_16702460 | 7.31 |
ENSRNOT00000061432
|
Ptk7
|
protein tyrosine kinase 7 |
| chrX_+_108287068 | 7.31 |
ENSRNOT00000093348
|
Il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chr15_+_45712821 | 7.31 |
ENSRNOT00000083381
ENSRNOT00000045833 |
Fam124a
|
family with sequence similarity 124 member A |
| chr7_-_130176696 | 7.30 |
ENSRNOT00000051347
|
Dennd6b
|
DENN domain containing 6B |
| chr6_-_46631983 | 7.30 |
ENSRNOT00000045963
|
Sox11
|
SRY box 11 |
| chr19_+_37652969 | 7.21 |
ENSRNOT00000041970
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr3_-_82734520 | 7.14 |
ENSRNOT00000083743
ENSRNOT00000012161 |
Ext2
|
exostosin glycosyltransferase 2 |
| chr1_+_105113595 | 7.13 |
ENSRNOT00000020853
ENSRNOT00000079655 |
Prmt3
|
protein arginine methyltransferase 3 |
| chr12_+_28212333 | 7.13 |
ENSRNOT00000001182
|
Auts2
|
autism susceptibility candidate 2 |
| chr6_-_33507542 | 7.10 |
ENSRNOT00000007928
|
Gdf7
|
growth differentiation factor 7 |
| chr7_+_54031316 | 7.06 |
ENSRNOT00000039096
|
Bbs10
|
Bardet-Biedl syndrome 10 |
| chr6_-_124735741 | 7.03 |
ENSRNOT00000064716
ENSRNOT00000091693 |
Rps6ka5
|
ribosomal protein S6 kinase A5 |
| chr8_-_32000378 | 7.01 |
ENSRNOT00000013341
|
Adamts15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ctcfl_Ctcf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.0 | 43.9 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 9.0 | 45.0 | GO:0061743 | motor learning(GO:0061743) |
| 8.6 | 59.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 7.5 | 59.8 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 7.0 | 55.9 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 7.0 | 20.9 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 6.3 | 18.8 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 6.1 | 49.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) regulation of cellular response to heat(GO:1900034) |
| 5.8 | 57.8 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 5.6 | 28.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 5.4 | 21.5 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 4.9 | 19.8 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 4.7 | 9.4 | GO:1900453 | negative regulation of long term synaptic depression(GO:1900453) |
| 4.5 | 13.5 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 4.5 | 44.8 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 4.4 | 17.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 4.4 | 13.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 4.3 | 30.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 4.1 | 24.9 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 3.8 | 15.2 | GO:0070778 | malate-aspartate shuttle(GO:0043490) L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 3.8 | 30.2 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 3.7 | 7.5 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 3.7 | 25.9 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 3.7 | 11.0 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 3.6 | 10.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 3.5 | 10.6 | GO:0000189 | MAPK import into nucleus(GO:0000189) regulation of chromatin assembly(GO:0010847) positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 3.4 | 23.9 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 3.2 | 9.7 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 3.1 | 9.4 | GO:2001055 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 3.1 | 9.4 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 3.1 | 21.6 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 3.0 | 8.9 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 2.9 | 17.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 2.9 | 31.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 2.8 | 11.0 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 2.6 | 13.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 2.5 | 7.5 | GO:0015942 | formate metabolic process(GO:0015942) |
| 2.4 | 7.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 2.4 | 7.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 2.3 | 11.6 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 2.3 | 13.8 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 2.2 | 13.4 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 2.1 | 14.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 2.1 | 8.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 2.1 | 316.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 2.1 | 20.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 2.0 | 12.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.9 | 13.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 1.8 | 7.3 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 1.8 | 7.2 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 1.8 | 7.0 | GO:0043988 | histone H3-S10 phosphorylation(GO:0043987) histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 1.7 | 10.4 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 1.7 | 8.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 1.6 | 11.5 | GO:0015808 | L-alanine transport(GO:0015808) |
| 1.6 | 6.6 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.6 | 18.1 | GO:0030432 | peristalsis(GO:0030432) |
| 1.6 | 6.4 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.6 | 33.4 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 1.6 | 6.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 1.4 | 12.9 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 1.4 | 12.6 | GO:0099612 | protein localization to axon(GO:0099612) |
| 1.4 | 9.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.3 | 4.0 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 1.3 | 11.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 1.2 | 6.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 1.2 | 9.6 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 1.2 | 7.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 1.2 | 7.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 1.2 | 14.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 1.1 | 4.6 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 1.1 | 30.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 1.1 | 37.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 1.1 | 3.4 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 1.1 | 11.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.1 | 7.6 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 1.1 | 5.3 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.0 | 6.3 | GO:0098597 | vocal learning(GO:0042297) imitative learning(GO:0098596) observational learning(GO:0098597) |
| 1.0 | 8.3 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 1.0 | 5.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 1.0 | 4.9 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 1.0 | 19.4 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 1.0 | 5.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 1.0 | 12.4 | GO:0007614 | short-term memory(GO:0007614) |
| 0.9 | 3.7 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.9 | 3.6 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.9 | 5.4 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.9 | 44.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.9 | 6.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.9 | 3.5 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 0.8 | 13.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.8 | 3.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.8 | 2.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.8 | 7.7 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.8 | 10.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.8 | 10.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.7 | 11.8 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.7 | 10.7 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.7 | 6.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.7 | 8.4 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.7 | 7.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.7 | 1.3 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.7 | 12.7 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.7 | 7.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.7 | 5.3 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.7 | 5.3 | GO:0061193 | taste bud development(GO:0061193) |
| 0.6 | 23.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.6 | 5.8 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.6 | 2.5 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.6 | 16.4 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.6 | 1.7 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) negative regulation of Toll signaling pathway(GO:0045751) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.5 | 3.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.5 | 4.9 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.5 | 4.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.5 | 4.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.5 | 3.8 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.5 | 6.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.5 | 8.7 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.5 | 1.8 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.5 | 5.4 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 7.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 7.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.4 | 2.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.4 | 3.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.4 | 2.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.4 | 6.2 | GO:0043090 | amino acid import(GO:0043090) |
| 0.4 | 13.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.4 | 1.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.4 | 11.6 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.4 | 7.3 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.4 | 6.5 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.4 | 5.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.4 | 4.0 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.4 | 0.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.4 | 1.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 3.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 19.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.3 | 1.4 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.3 | 10.2 | GO:0060065 | uterus development(GO:0060065) |
| 0.3 | 2.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.3 | 4.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.3 | 1.3 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.3 | 8.7 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.3 | 28.4 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.3 | 10.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 4.5 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.3 | 5.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 3.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 3.5 | GO:0060746 | parental behavior(GO:0060746) |
| 0.3 | 1.5 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.3 | 3.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 2.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 8.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 12.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.3 | 0.8 | GO:0021626 | hindbrain maturation(GO:0021578) cerebellum maturation(GO:0021590) central nervous system maturation(GO:0021626) cerebellar cortex maturation(GO:0021699) |
| 0.3 | 0.5 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 5.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 4.3 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.2 | 0.7 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 0.2 | 5.6 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 5.0 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 0.9 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.2 | 8.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 2.0 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 2.8 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.2 | 3.5 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 24.9 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.2 | 10.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 1.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 5.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.4 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.2 | 80.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.2 | 13.0 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.2 | 2.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 1.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 6.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 1.8 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.2 | 5.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 4.8 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.2 | 9.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.2 | 0.8 | GO:0033625 | positive regulation of integrin activation(GO:0033625) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 1.6 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 15.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 4.8 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.1 | 2.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 12.1 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.1 | 4.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 4.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.5 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 17.1 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 7.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.9 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 0.4 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 | 0.4 | GO:0019541 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 0.1 | 2.7 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.1 | 7.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 10.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 8.0 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.1 | 1.5 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 11.5 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.1 | 1.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 3.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 3.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 2.8 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 4.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 4.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 9.7 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 3.9 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.1 | 6.2 | GO:0030198 | extracellular matrix organization(GO:0030198) |
| 0.1 | 2.9 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 1.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 8.3 | GO:0048812 | neuron projection morphogenesis(GO:0048812) |
| 0.0 | 4.0 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.4 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 2.7 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.3 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.4 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 3.4 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.8 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 1.5 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 3.5 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) |
| 0.0 | 0.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.3 | GO:0001510 | RNA methylation(GO:0001510) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.2 | 49.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 7.9 | 158.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 4.7 | 23.6 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 4.1 | 20.7 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 4.1 | 81.4 | GO:0043196 | varicosity(GO:0043196) |
| 3.6 | 10.7 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 3.1 | 12.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 3.1 | 28.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 3.0 | 45.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 3.0 | 44.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 3.0 | 47.8 | GO:0005883 | neurofilament(GO:0005883) |
| 2.9 | 14.7 | GO:0016939 | kinesin II complex(GO:0016939) |
| 2.9 | 44.1 | GO:0031045 | dense core granule(GO:0031045) |
| 2.8 | 47.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 2.5 | 17.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 2.3 | 73.6 | GO:0005921 | gap junction(GO:0005921) |
| 2.2 | 11.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 2.2 | 13.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 2.0 | 18.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 2.0 | 7.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.8 | 14.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 1.8 | 7.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 1.7 | 30.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.7 | 8.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 1.6 | 6.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.6 | 8.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.6 | 25.7 | GO:0032039 | integrator complex(GO:0032039) |
| 1.6 | 11.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.4 | 16.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.3 | 10.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 1.2 | 36.5 | GO:0030673 | axolemma(GO:0030673) |
| 1.2 | 9.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.2 | 47.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 1.1 | 6.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.1 | 5.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 1.1 | 10.6 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 1.0 | 12.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.8 | 14.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.7 | 7.2 | GO:0044352 | F-actin capping protein complex(GO:0008290) pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.7 | 12.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.7 | 2.6 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.6 | 24.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.6 | 1.8 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.5 | 9.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.5 | 8.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 16.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.5 | 1.5 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.5 | 108.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.5 | 2.8 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.5 | 6.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 2.7 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.4 | 6.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 10.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.4 | 33.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.4 | 8.4 | GO:0001741 | XY body(GO:0001741) |
| 0.4 | 7.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.4 | 2.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.4 | 3.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 4.2 | GO:0002177 | manchette(GO:0002177) |
| 0.4 | 3.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.4 | 10.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.4 | 3.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.4 | 17.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.3 | 74.0 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.3 | 365.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.3 | 3.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 3.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 2.0 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.3 | 1.4 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.3 | 4.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 2.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 13.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 28.3 | GO:0098793 | presynapse(GO:0098793) |
| 0.2 | 6.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 2.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 1.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 1.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 4.4 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 3.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 13.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 3.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.7 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 3.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 4.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 1.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 9.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 25.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 19.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 5.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 34.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.1 | 2.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 8.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 4.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 21.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 1.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 4.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 1.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 2.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.5 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 5.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 17.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 2.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.6 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 16.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 16.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 123.1 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.6 | 81.5 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 12.2 | 49.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 7.6 | 30.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 7.0 | 20.9 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 6.9 | 20.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 5.9 | 17.7 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 5.7 | 158.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 5.5 | 11.0 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 4.7 | 28.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 4.3 | 30.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 3.8 | 11.5 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 3.8 | 18.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 3.6 | 18.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 3.6 | 24.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 3.3 | 16.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 3.3 | 19.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 3.2 | 9.7 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 3.1 | 12.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 3.0 | 15.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 3.0 | 12.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 2.9 | 11.6 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 2.6 | 12.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 2.6 | 7.7 | GO:0033222 | xylose binding(GO:0033222) |
| 2.3 | 14.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 2.1 | 8.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 2.0 | 6.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 1.9 | 9.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.9 | 13.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.9 | 7.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.8 | 7.3 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 1.8 | 10.9 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 1.8 | 14.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.5 | 10.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 1.5 | 7.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.5 | 7.5 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 1.5 | 13.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 1.4 | 19.8 | GO:0051378 | serotonin binding(GO:0051378) |
| 1.4 | 5.6 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 1.4 | 16.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 1.3 | 4.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 1.3 | 10.6 | GO:0043495 | protein anchor(GO:0043495) |
| 1.3 | 16.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.3 | 29.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 1.2 | 8.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 1.2 | 7.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 1.1 | 4.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 1.1 | 29.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 1.0 | 4.2 | GO:0019002 | GMP binding(GO:0019002) |
| 1.0 | 4.0 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 1.0 | 17.7 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 1.0 | 6.8 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 1.0 | 13.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.9 | 4.6 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.9 | 33.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.9 | 5.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.9 | 6.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.9 | 41.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.8 | 5.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.7 | 3.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.7 | 17.7 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.7 | 30.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.7 | 7.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.7 | 20.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.7 | 5.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.7 | 4.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.7 | 5.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.7 | 2.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.6 | 6.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.6 | 7.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.6 | 3.8 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.6 | 2.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.6 | 2.5 | GO:0034618 | arginine-tRNA ligase activity(GO:0004814) arginine binding(GO:0034618) |
| 0.6 | 6.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.6 | 11.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.6 | 2.3 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.6 | 161.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.6 | 5.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 5.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.5 | 385.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.5 | 7.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.5 | 4.8 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.4 | 13.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 5.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.4 | 4.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.4 | 9.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.4 | 19.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.4 | 7.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.4 | 5.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.4 | 2.5 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.4 | 10.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.4 | 3.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.3 | 13.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 52.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.3 | 3.9 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.3 | 8.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 6.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 2.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 19.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 14.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 0.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 8.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.3 | 10.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 15.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.2 | 3.9 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.2 | 0.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.2 | 1.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 1.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 7.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 3.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 17.0 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.2 | 19.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 6.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 2.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 14.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 0.7 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 4.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 1.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 3.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 5.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 17.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 9.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 2.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 1.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 2.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 2.1 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.7 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 9.1 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.1 | 1.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 6.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 9.7 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.1 | 3.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 15.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 2.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.5 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 8.6 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 1.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 11.8 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 1.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.7 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 49.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.9 | 30.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 1.8 | 47.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 1.8 | 70.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.9 | 23.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.5 | 17.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.5 | 9.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.5 | 18.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.5 | 8.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 16.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.4 | 14.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.4 | 14.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.4 | 3.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 23.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.4 | 25.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.4 | 16.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.4 | 7.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 13.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.3 | 11.8 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.3 | 7.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 8.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 15.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 41.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 3.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 4.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 9.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 8.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 14.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.0 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.1 | 4.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 59.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 2.0 | 18.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 1.8 | 52.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.7 | 55.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.6 | 24.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.6 | 17.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 1.4 | 19.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.1 | 9.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 1.0 | 13.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.0 | 24.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 1.0 | 14.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.9 | 24.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.9 | 44.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.9 | 28.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.8 | 21.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.7 | 22.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.6 | 32.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.6 | 10.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.6 | 9.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.5 | 9.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.5 | 13.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.5 | 9.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.5 | 10.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.5 | 7.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 15.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 7.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.4 | 7.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.4 | 10.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.4 | 9.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.3 | 12.5 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.3 | 8.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 7.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 2.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.3 | 8.7 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.3 | 11.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 4.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 2.1 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.3 | 13.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 7.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 41.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 13.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 3.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 15.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 1.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 4.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 4.8 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 5.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 5.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 3.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 2.9 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |