Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
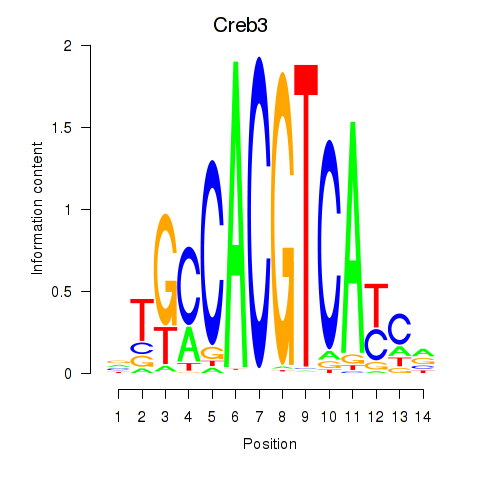
Results for Creb3
Z-value: 0.95
Transcription factors associated with Creb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Creb3
|
ENSRNOG00000016452 | cAMP responsive element binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3 | rn6_v1_chr5_+_59063531_59063558 | -0.12 | 3.5e-02 | Click! |
Activity profile of Creb3 motif
Sorted Z-values of Creb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_40247436 | 36.78 |
ENSRNOT00000079830
|
Gpx3
|
glutathione peroxidase 3 |
| chr3_-_175709465 | 33.30 |
ENSRNOT00000089971
|
Gata5
|
GATA binding protein 5 |
| chr7_+_141249044 | 33.19 |
ENSRNOT00000084911
|
Aqp5
|
aquaporin 5 |
| chr7_+_120749752 | 29.94 |
ENSRNOT00000087888
|
Kdelr3
|
KDEL endoplasmic reticulum protein retention receptor 3 |
| chr8_+_58431407 | 28.75 |
ENSRNOT00000011974
|
Sln
|
sarcolipin |
| chr2_-_257474733 | 24.46 |
ENSRNOT00000016967
ENSRNOT00000066780 |
Nexn
|
nexilin (F actin binding protein) |
| chr1_-_141893674 | 23.99 |
ENSRNOT00000019059
ENSRNOT00000085988 |
Idh2
|
isocitrate dehydrogenase (NADP(+)) 2, mitochondrial |
| chr8_-_25904564 | 22.84 |
ENSRNOT00000082744
ENSRNOT00000064783 |
Tbx20
|
T-box 20 |
| chr19_-_38120578 | 20.68 |
ENSRNOT00000026873
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr1_-_101085884 | 19.89 |
ENSRNOT00000085352
|
Rcn3
|
reticulocalbin 3 |
| chr2_+_227455722 | 19.38 |
ENSRNOT00000064809
|
Sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr8_-_21995806 | 18.77 |
ENSRNOT00000028034
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr8_-_13109487 | 17.15 |
ENSRNOT00000061616
|
Amotl1
|
angiomotin-like 1 |
| chr3_-_146470293 | 15.49 |
ENSRNOT00000009627
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr10_+_36099263 | 14.86 |
ENSRNOT00000083568
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr5_-_173081839 | 14.15 |
ENSRNOT00000066880
|
Mmp23
|
matrix metallopeptidase 23 |
| chr10_-_62483342 | 13.66 |
ENSRNOT00000079956
|
LOC100912068
|
hypermethylated in cancer 1 protein-like |
| chr18_-_28428117 | 13.55 |
ENSRNOT00000078202
|
Slc23a1
|
solute carrier family 23 member 1 |
| chr6_-_7741414 | 13.33 |
ENSRNOT00000038246
|
Thada
|
THADA, armadillo repeat containing |
| chr10_+_16970626 | 13.30 |
ENSRNOT00000005383
|
Dusp1
|
dual specificity phosphatase 1 |
| chr3_+_95232166 | 12.91 |
ENSRNOT00000017952
|
LOC691083
|
hypothetical protein LOC691083 |
| chr1_-_87174530 | 12.73 |
ENSRNOT00000089578
|
Kcnk6
|
potassium two pore domain channel subfamily K member 6 |
| chr1_-_101086198 | 12.72 |
ENSRNOT00000027917
|
Rcn3
|
reticulocalbin 3 |
| chr1_-_88176610 | 12.48 |
ENSRNOT00000032394
|
LOC100909725
|
potassium channel subfamily K member 6-like |
| chr10_-_70337532 | 11.80 |
ENSRNOT00000055963
|
Slfn13
|
schlafen family member 13 |
| chr8_+_117117430 | 11.71 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr3_+_45538797 | 11.65 |
ENSRNOT00000007632
|
Dapl1
|
death associated protein-like 1 |
| chr4_+_98648545 | 11.46 |
ENSRNOT00000008451
|
Eif2ak3
|
eukaryotic translation initiation factor 2 alpha kinase 3 |
| chrX_+_156999826 | 11.42 |
ENSRNOT00000081819
|
Idh3g
|
isocitrate dehydrogenase 3 (NAD), gamma |
| chr15_+_57891680 | 11.01 |
ENSRNOT00000001383
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr7_+_114724610 | 11.01 |
ENSRNOT00000014541
|
Dennd3
|
DENN domain containing 3 |
| chr15_-_61564695 | 10.99 |
ENSRNOT00000068216
|
Rgcc
|
regulator of cell cycle |
| chr20_+_44680449 | 10.71 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chrX_-_156399760 | 10.67 |
ENSRNOT00000086921
|
Fam50a
|
family with sequence similarity 50, member A |
| chr5_+_164845925 | 9.81 |
ENSRNOT00000011384
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr4_+_56625561 | 9.69 |
ENSRNOT00000008356
ENSRNOT00000090808 ENSRNOT00000008972 |
Calu
|
calumenin |
| chr8_-_116361343 | 9.52 |
ENSRNOT00000066296
|
Sema3b
|
semaphorin 3B |
| chr18_+_52423883 | 9.49 |
ENSRNOT00000022017
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr10_-_65805693 | 9.43 |
ENSRNOT00000012245
|
Tnfaip1
|
TNF alpha induced protein 1 |
| chr2_-_33841499 | 9.39 |
ENSRNOT00000040533
|
Srek1
|
splicing regulatory glutamic acid and lysine rich protein 1 |
| chr2_-_123282214 | 9.13 |
ENSRNOT00000021156
|
Ccna2
|
cyclin A2 |
| chr11_+_46009322 | 9.07 |
ENSRNOT00000002233
|
Tmem45a
|
transmembrane protein 45A |
| chr13_-_61070599 | 8.93 |
ENSRNOT00000005251
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr11_+_46714355 | 8.86 |
ENSRNOT00000073404
|
Tmem45al
|
transmembrane protein 45A-like |
| chr1_-_82120902 | 8.84 |
ENSRNOT00000027684
|
Erf
|
Ets2 repressor factor |
| chr10_-_40764185 | 8.80 |
ENSRNOT00000017486
|
Sparc
|
secreted protein acidic and cysteine rich |
| chr1_+_101859346 | 8.75 |
ENSRNOT00000028627
|
Kdelr1
|
KDEL endoplasmic reticulum protein retention receptor 1 |
| chr2_-_157759819 | 8.63 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr1_-_216663720 | 8.51 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr1_+_144831523 | 8.43 |
ENSRNOT00000039748
|
Mex3b
|
mex-3 RNA binding family member B |
| chr8_-_49308806 | 8.42 |
ENSRNOT00000047291
|
Cd3e
|
CD3e molecule |
| chr1_+_199941161 | 8.35 |
ENSRNOT00000027616
|
Bag3
|
Bcl2-associated athanogene 3 |
| chr19_-_37210412 | 8.28 |
ENSRNOT00000083097
|
B3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr15_+_45422010 | 7.94 |
ENSRNOT00000012231
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr7_-_2819299 | 7.85 |
ENSRNOT00000045547
|
Slc39a5
|
solute carrier family 39 member 5 |
| chr17_+_43616247 | 7.83 |
ENSRNOT00000072019
|
Hist1h3a
|
histone cluster 1, H3a |
| chr12_+_7081895 | 7.73 |
ENSRNOT00000047163
|
Hmgb1
|
high mobility group box 1 |
| chr20_+_46250363 | 7.71 |
ENSRNOT00000076522
ENSRNOT00000000334 |
Cd164
|
CD164 molecule |
| chr6_+_132702448 | 7.63 |
ENSRNOT00000005743
|
Yy1
|
YY1 transcription factor |
| chr15_-_34322115 | 7.50 |
ENSRNOT00000026734
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr3_+_150801289 | 7.47 |
ENSRNOT00000035060
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr2_-_123281856 | 7.23 |
ENSRNOT00000079745
|
Ccna2
|
cyclin A2 |
| chr2_+_233615739 | 7.14 |
ENSRNOT00000051009
|
Pitx2
|
paired-like homeodomain 2 |
| chrX_-_156999650 | 7.08 |
ENSRNOT00000083557
|
Ssr4
|
signal sequence receptor subunit 4 |
| chr17_-_1797732 | 6.96 |
ENSRNOT00000033017
ENSRNOT00000079742 |
Cdc14b
|
cell division cycle 14B |
| chr14_-_19072677 | 6.92 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr17_-_15718015 | 6.73 |
ENSRNOT00000048653
ENSRNOT00000090647 |
Bicd2
|
BICD cargo adaptor 2 |
| chr5_+_63192298 | 6.21 |
ENSRNOT00000008329
|
Sec61b
|
Sec61 translocon beta subunit |
| chr5_-_138470096 | 6.11 |
ENSRNOT00000011392
|
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr3_+_113415774 | 6.08 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr6_-_91456696 | 5.99 |
ENSRNOT00000005577
|
Rps29
|
ribosomal protein S29 |
| chr10_+_37032043 | 5.98 |
ENSRNOT00000037303
ENSRNOT00000091242 |
Phykpl
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr1_+_167309051 | 5.80 |
ENSRNOT00000055235
ENSRNOT00000076894 |
Pgap2
|
post-GPI attachment to proteins 2 |
| chr3_-_122920684 | 5.72 |
ENSRNOT00000028818
|
Cpxm1
|
carboxypeptidase X (M14 family), member 1 |
| chrX_+_78259409 | 5.70 |
ENSRNOT00000049779
|
RGD1560455
|
similar to RIKEN cDNA A630033H20 gene |
| chr2_-_20152884 | 5.70 |
ENSRNOT00000021941
|
Atg10
|
autophagy related 10 |
| chr11_-_64752544 | 5.63 |
ENSRNOT00000048738
|
Tmem39a
|
transmembrane protein 39a |
| chr10_+_34938667 | 5.54 |
ENSRNOT00000086443
ENSRNOT00000059104 |
LOC103690164
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr4_-_57625147 | 5.53 |
ENSRNOT00000074649
ENSRNOT00000078961 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr10_+_56610051 | 5.50 |
ENSRNOT00000024348
|
Dvl2
|
dishevelled segment polarity protein 2 |
| chr19_-_37938857 | 5.34 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr1_-_87174821 | 5.29 |
ENSRNOT00000027974
|
Kcnk6
|
potassium two pore domain channel subfamily K member 6 |
| chr10_+_56524468 | 5.20 |
ENSRNOT00000022041
|
Gps2
|
G protein pathway suppressor 2 |
| chr14_+_11199404 | 5.20 |
ENSRNOT00000003106
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chrX_-_42329232 | 4.87 |
ENSRNOT00000045705
|
LOC100361934
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 |
| chr8_-_96203677 | 4.83 |
ENSRNOT00000074515
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr11_-_67647818 | 4.81 |
ENSRNOT00000003068
|
Wdr5b
|
WD repeat domain 5B |
| chr1_-_282170017 | 4.73 |
ENSRNOT00000066947
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr1_+_80141630 | 4.69 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr6_+_10912383 | 4.52 |
ENSRNOT00000061747
ENSRNOT00000086247 |
Ttc7a
|
tetratricopeptide repeat domain 7A |
| chr1_+_140601791 | 4.51 |
ENSRNOT00000091588
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr5_-_117612123 | 4.47 |
ENSRNOT00000065112
|
Dock7
|
dedicator of cytokinesis 7 |
| chr4_-_66002444 | 4.43 |
ENSRNOT00000018645
|
Zc3hav1l
|
zinc finger CCCH-type containing, antiviral 1 like |
| chr14_-_79988110 | 4.23 |
ENSRNOT00000090597
|
Afap1
|
actin filament associated protein 1 |
| chr1_+_114046478 | 4.19 |
ENSRNOT00000032254
|
Siglech
|
sialic acid binding Ig-like lectin H |
| chr17_+_10463303 | 4.18 |
ENSRNOT00000060822
|
Rnf44
|
ring finger protein 44 |
| chr20_+_31298269 | 4.18 |
ENSRNOT00000000673
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr8_-_39243705 | 4.14 |
ENSRNOT00000043518
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr9_-_17827032 | 4.05 |
ENSRNOT00000026929
|
Slc35b2
|
solute carrier family 35 member B2 |
| chr7_-_59547174 | 3.95 |
ENSRNOT00000085836
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr17_+_89742140 | 3.94 |
ENSRNOT00000077899
|
Mastl
|
microtubule associated serine/threonine kinase-like |
| chr17_+_26925828 | 3.91 |
ENSRNOT00000018310
|
Txndc5
|
thioredoxin domain containing 5 |
| chr1_-_167308827 | 3.77 |
ENSRNOT00000027590
ENSRNOT00000027575 |
Nup98
|
nucleoporin 98 |
| chr10_-_75120247 | 3.77 |
ENSRNOT00000011402
|
Lpo
|
lactoperoxidase |
| chr12_+_9728486 | 3.76 |
ENSRNOT00000001263
|
Lnx2
|
ligand of numb-protein X 2 |
| chr3_+_177351518 | 3.65 |
ENSRNOT00000023989
|
Pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr14_+_11198896 | 3.63 |
ENSRNOT00000079767
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr5_+_160306727 | 3.63 |
ENSRNOT00000016648
|
Agmat
|
agmatinase |
| chr3_+_146695344 | 3.62 |
ENSRNOT00000010955
|
Gins1
|
GINS complex subunit 1 |
| chr14_+_35683442 | 3.44 |
ENSRNOT00000003085
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr10_+_4578469 | 3.43 |
ENSRNOT00000003332
|
Txndc11
|
thioredoxin domain containing 11 |
| chr14_+_7949239 | 3.42 |
ENSRNOT00000044617
|
Ndufb4l1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4-like 1 |
| chr13_+_89253617 | 3.40 |
ENSRNOT00000044575
|
LOC100362384
|
ferritin light chain 1-like |
| chr16_-_71125316 | 3.35 |
ENSRNOT00000020900
|
Plpp5
|
phospholipid phosphatase 5 |
| chr15_-_34338956 | 3.32 |
ENSRNOT00000026914
|
Mdp1
|
magnesium-dependent phosphatase 1 |
| chr12_-_23021517 | 3.18 |
ENSRNOT00000041654
|
Myl10
|
myosin light chain 10 |
| chr3_+_67538289 | 3.08 |
ENSRNOT00000009839
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr16_+_2338333 | 3.08 |
ENSRNOT00000017692
|
Arf4
|
ADP-ribosylation factor 4 |
| chr11_-_66819079 | 3.06 |
ENSRNOT00000003255
|
Golgb1
|
golgin B1 |
| chr10_-_103736973 | 2.89 |
ENSRNOT00000004372
|
Nat9
|
N-acetyltransferase 9 |
| chr12_-_19314016 | 2.87 |
ENSRNOT00000001825
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr7_+_2737765 | 2.86 |
ENSRNOT00000004895
|
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr14_-_79987915 | 2.85 |
ENSRNOT00000089006
|
Afap1
|
actin filament associated protein 1 |
| chr12_-_6630274 | 2.82 |
ENSRNOT00000087840
|
AABR07035194.1
|
|
| chr16_-_49484985 | 2.82 |
ENSRNOT00000016723
|
Ufsp2
|
UFM1-specific peptidase 2 |
| chr9_+_55050203 | 2.82 |
ENSRNOT00000021403
|
Nabp1
|
nucleic acid binding protein 1 |
| chr8_+_59164572 | 2.77 |
ENSRNOT00000015102
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr8_-_39243882 | 2.77 |
ENSRNOT00000082086
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr3_+_8643936 | 2.75 |
ENSRNOT00000077797
|
Set
|
SET nuclear proto-oncogene |
| chr4_-_165118062 | 2.74 |
ENSRNOT00000087219
|
LOC502908
|
Ly49s8 |
| chr1_+_221420271 | 2.72 |
ENSRNOT00000028481
|
LOC687780
|
similar to Finkel-Biskis-Reilly murine sarcoma virusubiquitously expressed |
| chr4_+_119997268 | 2.71 |
ENSRNOT00000073429
|
Rpn1
|
ribophorin I |
| chr3_-_94686989 | 2.64 |
ENSRNOT00000016677
|
Depdc7
|
DEP domain containing 7 |
| chr9_-_100592271 | 2.55 |
ENSRNOT00000086433
|
Hdlbp
|
high density lipoprotein binding protein |
| chr20_+_3611964 | 2.50 |
ENSRNOT00000090510
ENSRNOT00000068159 |
Dpcr1
|
diffuse panbronchiolitis critical region 1 |
| chrX_+_120860178 | 2.48 |
ENSRNOT00000088661
|
Wdr44
|
WD repeat domain 44 |
| chr5_+_5215196 | 2.45 |
ENSRNOT00000010481
|
Tram1
|
translocation associated membrane protein 1 |
| chr9_+_10013854 | 2.42 |
ENSRNOT00000077653
ENSRNOT00000072033 |
Khsrp
|
KH-type splicing regulatory protein |
| chr3_+_138504214 | 2.41 |
ENSRNOT00000091529
|
Kat14
|
lysine acetyltransferase 14 |
| chrX_-_123474154 | 2.38 |
ENSRNOT00000092415
ENSRNOT00000092455 |
RGD1564541
|
similar to hypothetical protein FLJ22965 |
| chr13_+_35554964 | 2.35 |
ENSRNOT00000072632
|
Tmem185b
|
transmembrane protein 185B |
| chr11_+_65960277 | 2.33 |
ENSRNOT00000003674
|
Ndufb4
|
NADH:ubiquinone oxidoreductase subunit B4 |
| chr1_-_100865894 | 2.22 |
ENSRNOT00000027580
|
Ptov1
|
prostate tumor overexpressed 1 |
| chr19_-_34105610 | 2.18 |
ENSRNOT00000017347
|
Prmt9
|
protein arginine methyltransferase 9 |
| chr11_-_71591502 | 2.17 |
ENSRNOT00000002403
|
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha |
| chrX_+_120859968 | 1.96 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr17_-_84614228 | 1.86 |
ENSRNOT00000043042
|
AABR07028748.1
|
|
| chr1_-_84008293 | 1.86 |
ENSRNOT00000002053
|
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr5_-_147846095 | 1.85 |
ENSRNOT00000072165
|
Txlna
|
taxilin alpha |
| chr13_-_88497901 | 1.70 |
ENSRNOT00000058560
|
Uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr5_-_152247332 | 1.68 |
ENSRNOT00000077165
|
Lin28a
|
lin-28 homolog A |
| chr1_-_183729569 | 1.64 |
ENSRNOT00000083293
|
Copb1
|
coatomer protein complex, subunit beta 1 |
| chr1_+_256370850 | 1.63 |
ENSRNOT00000030962
|
Cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr7_-_11406771 | 1.63 |
ENSRNOT00000047450
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr19_-_37990353 | 1.55 |
ENSRNOT00000026817
|
Ddx28
|
DEAD-box helicase 28 |
| chr10_+_82375572 | 1.52 |
ENSRNOT00000004947
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr8_+_81863619 | 1.50 |
ENSRNOT00000080608
|
Fam214a
|
family with sequence similarity 214, member A |
| chr6_+_91476698 | 1.47 |
ENSRNOT00000005608
|
Mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr10_+_103737162 | 1.47 |
ENSRNOT00000055037
|
Tmem104
|
transmembrane protein 104 |
| chr10_+_38692211 | 1.45 |
ENSRNOT00000009440
|
Aff4
|
AF4/FMR2 family, member 4 |
| chr2_-_231881939 | 1.44 |
ENSRNOT00000074644
|
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr2_-_2891253 | 1.43 |
ENSRNOT00000029250
|
Arsk
|
arylsulfatase family, member K |
| chr7_-_2972521 | 1.35 |
ENSRNOT00000061995
|
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr18_+_1504178 | 1.35 |
ENSRNOT00000048198
|
Rpl36al
|
ribosomal protein L36a-like |
| chr1_-_205842428 | 1.34 |
ENSRNOT00000024655
|
Dhx32
|
DEAH-box helicase 32 (putative) |
| chr8_-_50539331 | 1.26 |
ENSRNOT00000088997
|
AABR07073400.1
|
|
| chr8_-_112594261 | 1.24 |
ENSRNOT00000015240
|
Uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr2_+_195996521 | 1.14 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chr5_+_76150840 | 1.13 |
ENSRNOT00000020702
|
Dnajc25
|
DnaJ heat shock protein family (Hsp40) member C25 |
| chr15_-_39893979 | 1.10 |
ENSRNOT00000068353
|
Cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr10_-_105149971 | 1.08 |
ENSRNOT00000012436
|
Srp68
|
signal recognition particle 68 |
| chr3_-_124461332 | 1.07 |
ENSRNOT00000073088
|
LOC103691893
|
putative ERV-F(c)1 provirus ancestral Env polyprotein |
| chr9_-_100624638 | 1.06 |
ENSRNOT00000051155
|
Hdlbp
|
high density lipoprotein binding protein |
| chr7_-_123287721 | 1.03 |
ENSRNOT00000009152
|
LOC100359574
|
NHP2 non-histone chromosome protein 2-like 1-like |
| chr6_-_26875376 | 1.01 |
ENSRNOT00000011838
|
Tmem214
|
transmembrane protein 214 |
| chr16_+_61758917 | 0.99 |
ENSRNOT00000084858
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr2_-_261402880 | 0.99 |
ENSRNOT00000052396
ENSRNOT00000077839 ENSRNOT00000088561 |
Fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr8_-_104995725 | 0.97 |
ENSRNOT00000037120
|
Slc25a36
|
solute carrier family 25 member 36 |
| chr1_-_219450451 | 0.94 |
ENSRNOT00000025317
|
Rad9a
|
RAD9 checkpoint clamp component A |
| chr10_-_96584896 | 0.94 |
ENSRNOT00000004699
ENSRNOT00000055073 |
Prkca
|
protein kinase C, alpha |
| chr10_-_3321404 | 0.92 |
ENSRNOT00000093722
ENSRNOT00000003284 |
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr18_-_56534415 | 0.92 |
ENSRNOT00000024374
|
Slc26a2
|
solute carrier family 26 member 2 |
| chr9_+_44491177 | 0.89 |
ENSRNOT00000075242
|
NEWGENE_1308196
|
mitochondrial ribosomal protein L30 |
| chr10_-_105228547 | 0.89 |
ENSRNOT00000028875
|
Srp68
|
signal recognition particle 68 |
| chr4_+_7260575 | 0.77 |
ENSRNOT00000064893
|
Fastk
|
Fas-activated serine/threonine kinase |
| chr1_-_221420115 | 0.69 |
ENSRNOT00000028475
|
Mrpl49
|
mitochondrial ribosomal protein L49 |
| chr1_-_220067123 | 0.69 |
ENSRNOT00000071020
ENSRNOT00000072373 |
Rbm14
|
RNA binding motif protein 14 |
| chr9_-_81566642 | 0.58 |
ENSRNOT00000080345
|
Aamp
|
angio-associated, migratory cell protein |
| chr3_-_79892429 | 0.58 |
ENSRNOT00000016191
|
Slc39a13
|
solute carrier family 39 member 13 |
| chr2_-_30846149 | 0.57 |
ENSRNOT00000025506
|
Slc30a5
|
solute carrier family 30 member 5 |
| chr7_-_116872511 | 0.56 |
ENSRNOT00000010120
|
Zc3h3
|
zinc finger CCCH type containing 3 |
| chr2_-_152824547 | 0.54 |
ENSRNOT00000019824
|
Dhx36
|
DEAH-box helicase 36 |
| chr4_+_55772377 | 0.48 |
ENSRNOT00000047698
|
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr5_+_155935554 | 0.45 |
ENSRNOT00000031855
|
LOC100911993
|
ubiquitin carboxyl-terminal hydrolase 48-like |
| chr3_+_160467552 | 0.45 |
ENSRNOT00000066657
|
Stk4
|
serine/threonine kinase 4 |
| chr7_+_72772440 | 0.44 |
ENSRNOT00000009989
|
Mtdh
|
metadherin |
| chr8_+_5790034 | 0.40 |
ENSRNOT00000061887
|
Mmp27
|
matrix metallopeptidase 27 |
| chr5_-_59198650 | 0.40 |
ENSRNOT00000020958
|
Olr833
|
olfactory receptor 833 |
| chr20_+_28027054 | 0.39 |
ENSRNOT00000071386
ENSRNOT00000001044 |
Ranbp2
|
RAN binding protein 2 |
| chr4_-_84768249 | 0.35 |
ENSRNOT00000013205
|
Fkbp14
|
FK506 binding protein 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Creb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 30.0 | GO:0060577 | pulmonary vein morphogenesis(GO:0060577) |
| 6.4 | 38.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 5.7 | 17.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 5.5 | 38.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 5.3 | 36.8 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 5.2 | 15.5 | GO:0019541 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 4.7 | 33.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 4.5 | 13.6 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 4.1 | 28.7 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 3.9 | 11.7 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 3.8 | 11.5 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 3.7 | 11.0 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 3.7 | 11.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 3.1 | 18.8 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 2.7 | 16.4 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 2.6 | 7.8 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 2.6 | 7.7 | GO:0035711 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) regulation of mismatch repair(GO:0032423) positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) T-helper 1 cell activation(GO:0035711) positive regulation of glycogen catabolic process(GO:0045819) negative regulation of apoptotic cell clearance(GO:2000426) |
| 2.6 | 18.0 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 2.1 | 6.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 2.0 | 24.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.9 | 7.6 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 1.7 | 8.4 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 1.6 | 9.8 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 1.6 | 10.9 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 1.5 | 4.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.5 | 8.9 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 1.4 | 8.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 1.4 | 8.4 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.3 | 3.8 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 1.2 | 33.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 1.2 | 4.7 | GO:0002188 | translation reinitiation(GO:0002188) |
| 1.1 | 8.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 1.0 | 13.3 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 1.0 | 6.9 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 1.0 | 2.9 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.9 | 5.5 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.8 | 4.8 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.7 | 8.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.7 | 13.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.7 | 4.0 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.7 | 20.7 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.6 | 4.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.6 | 6.7 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.6 | 9.7 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.6 | 3.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.5 | 1.6 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.5 | 10.7 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.5 | 5.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.5 | 4.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.5 | 7.5 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.5 | 8.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.5 | 8.8 | GO:0009629 | response to gravity(GO:0009629) response to L-ascorbic acid(GO:0033591) |
| 0.5 | 3.6 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.4 | 7.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.4 | 3.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.4 | 9.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.4 | 1.6 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.4 | 1.5 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.3 | 2.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.3 | 6.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.3 | 1.7 | GO:0010587 | miRNA catabolic process(GO:0010587) positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.3 | 0.9 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.3 | 14.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 3.4 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 5.8 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.2 | 2.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 5.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 0.5 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.2 | 4.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.2 | 4.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 5.2 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 1.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.0 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 8.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 3.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.9 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.1 | 7.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 4.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 1.0 | GO:1901679 | nucleotide transmembrane transport(GO:1901679) |
| 0.1 | 6.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.4 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 1.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 3.6 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.1 | 0.4 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 9.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.6 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 2.8 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.1 | 0.6 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 0.9 | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902229) |
| 0.1 | 7.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 11.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 0.2 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.1 | 5.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 6.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 3.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 2.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 1.8 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 2.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 3.6 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 2.5 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 2.8 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.6 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) |
| 0.0 | 4.1 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 3.1 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.4 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.0 | 1.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.4 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 4.7 | 14.2 | GO:0005962 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 2.8 | 11.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 2.7 | 13.3 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 2.4 | 9.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 2.2 | 8.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 2.1 | 10.6 | GO:0097413 | Lewy body(GO:0097413) |
| 2.0 | 7.9 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.2 | 3.6 | GO:0000811 | GINS complex(GO:0000811) |
| 1.2 | 16.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 1.2 | 7.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 1.1 | 8.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.9 | 3.8 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.8 | 28.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.8 | 46.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.8 | 4.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.7 | 2.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.6 | 4.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) GAIT complex(GO:0097452) |
| 0.6 | 4.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.6 | 9.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.6 | 15.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 17.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.4 | 7.6 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.4 | 18.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 2.0 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.3 | 6.8 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.3 | 3.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 2.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 2.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 8.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 24.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 0.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 1.0 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 12.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 9.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.9 | GO:0019028 | viral capsid(GO:0019028) |
| 0.1 | 2.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 4.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 7.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 29.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 6.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 10.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 8.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 7.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 10.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 20.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 2.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 11.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 23.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 47.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 11.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 9.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 4.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 10.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 17.3 | GO:0005654 | nucleoplasm(GO:0005654) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 38.7 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 6.4 | 38.2 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 4.6 | 36.8 | GO:0008430 | selenium binding(GO:0008430) |
| 4.5 | 13.6 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 4.4 | 13.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 3.9 | 15.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 2.4 | 33.2 | GO:0015250 | water channel activity(GO:0015250) |
| 1.9 | 11.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.9 | 18.8 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 1.6 | 7.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.6 | 6.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 1.5 | 7.7 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 1.2 | 6.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 1.0 | 11.2 | GO:0070513 | death domain binding(GO:0070513) |
| 1.0 | 4.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.9 | 1.9 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.9 | 3.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.9 | 4.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.9 | 13.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.8 | 6.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.8 | 4.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.8 | 8.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.7 | 7.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.7 | 9.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.7 | 30.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.6 | 8.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.6 | 11.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.5 | 1.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.5 | 24.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 16.4 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.5 | 2.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.5 | 9.8 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.5 | 8.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.4 | 5.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.4 | 2.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 8.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 11.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.4 | 40.9 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.4 | 4.0 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.4 | 7.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 1.0 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 0.3 | 0.9 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.3 | 9.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 3.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.3 | 5.9 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.3 | 3.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 7.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 11.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 29.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 8.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.2 | 8.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 1.7 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 5.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 10.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 11.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.2 | 3.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 4.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 2.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 3.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 3.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.2 | 7.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 8.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 7.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 3.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 18.0 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.1 | 23.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 1.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.0 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 14.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 3.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 3.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 8.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.8 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 5.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 13.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.5 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 30.6 | GO:0030234 | enzyme regulator activity(GO:0030234) |
| 0.0 | 3.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 2.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 24.5 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 5.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 16.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.8 | 18.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.6 | 33.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.5 | 15.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 7.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 18.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 13.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 5.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 7.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 8.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 7.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 3.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 8.9 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 11.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 15.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 9.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 3.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 8.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 33.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 2.1 | 38.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 1.9 | 15.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.4 | 18.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.2 | 11.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.1 | 19.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 1.1 | 8.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.0 | 16.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 1.0 | 23.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.8 | 1.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.8 | 7.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.6 | 6.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.5 | 29.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.4 | 8.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.4 | 12.8 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.4 | 17.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.3 | 41.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.3 | 14.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 10.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 19.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 16.0 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.2 | 8.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 4.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 2.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 4.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 25.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 4.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 1.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 6.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 2.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |