Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
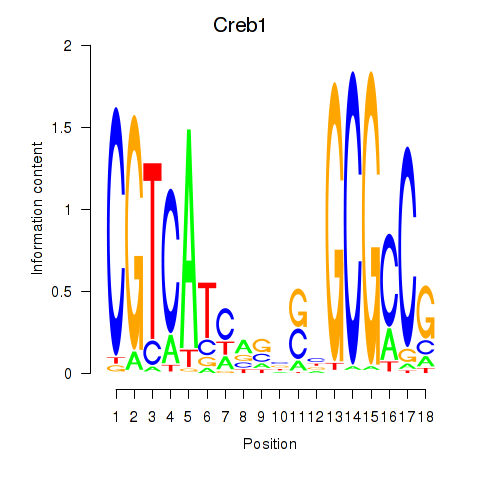
Results for Creb1
Z-value: 1.32
Transcription factors associated with Creb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Creb1
|
ENSRNOG00000013412 | cAMP responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb1 | rn6_v1_chr9_+_71230108_71230108 | 0.47 | 3.7e-19 | Click! |
Activity profile of Creb1 motif
Sorted Z-values of Creb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_132032944 | 30.09 |
ENSRNOT00000089278
|
Kif15
|
kinesin family member 15 |
| chr9_-_10757720 | 22.03 |
ENSRNOT00000083848
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr1_-_219863926 | 18.70 |
ENSRNOT00000026454
ENSRNOT00000066455 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr12_+_504007 | 18.37 |
ENSRNOT00000001475
|
Brca2
|
BRCA2, DNA repair associated |
| chr1_+_221448661 | 17.92 |
ENSRNOT00000072493
|
LOC100910252
|
sororin-like |
| chrX_+_105147153 | 17.49 |
ENSRNOT00000088172
|
Cenpi
|
centromere protein I |
| chr19_-_56731372 | 17.12 |
ENSRNOT00000024182
|
Nup133
|
nucleoporin 133 |
| chr7_-_28040510 | 16.95 |
ENSRNOT00000005674
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr6_+_144156175 | 16.55 |
ENSRNOT00000006582
|
Esyt2
|
extended synaptotagmin 2 |
| chr6_-_27080856 | 16.46 |
ENSRNOT00000012512
|
Cenpa
|
centromere protein A |
| chr17_+_18358712 | 16.06 |
ENSRNOT00000001979
|
Nup153
|
nucleoporin 153 |
| chr10_-_104482838 | 15.81 |
ENSRNOT00000007246
|
Recql5
|
RecQ like helicase 5 |
| chr1_+_29915443 | 15.09 |
ENSRNOT00000065677
|
Cenpw
|
centromere protein W |
| chr3_-_150073721 | 14.61 |
ENSRNOT00000022428
|
E2f1
|
E2F transcription factor 1 |
| chr1_-_219412816 | 14.57 |
ENSRNOT00000083204
ENSRNOT00000029580 |
Rps6kb2
|
ribosomal protein S6 kinase B2 |
| chr10_-_85725429 | 13.25 |
ENSRNOT00000005471
|
Rpl23
|
ribosomal protein L23 |
| chr10_-_103826448 | 12.82 |
ENSRNOT00000085636
|
Fdxr
|
ferredoxin reductase |
| chr1_+_220325352 | 12.18 |
ENSRNOT00000027258
|
LOC108348098
|
breast cancer metastasis-suppressor 1 homolog |
| chr14_+_1469748 | 12.14 |
ENSRNOT00000071648
|
Ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr1_-_198450688 | 12.02 |
ENSRNOT00000027392
|
Pagr1
|
Paxip1-associated glutamate-rich protein 1 |
| chr15_-_24056073 | 11.99 |
ENSRNOT00000015100
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr6_+_52751106 | 11.94 |
ENSRNOT00000014373
|
Twistnb
|
TWIST neighbor |
| chr7_-_70452675 | 11.52 |
ENSRNOT00000090498
|
AC114111.1
|
|
| chr7_-_24004774 | 11.43 |
ENSRNOT00000007549
|
Pwp1
|
PWP1 homolog, endonuclein |
| chr7_+_70452579 | 11.38 |
ENSRNOT00000046099
|
B4galnt1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr3_-_176791960 | 11.31 |
ENSRNOT00000018237
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr14_+_44524416 | 10.83 |
ENSRNOT00000038927
|
Rpl9
|
ribosomal protein L9 |
| chr15_+_34431357 | 10.83 |
ENSRNOT00000027604
|
Nop9
|
NOP9 nucleolar protein |
| chr8_-_49271834 | 10.80 |
ENSRNOT00000085022
|
Ube4a
|
ubiquitination factor E4A |
| chr6_+_51662224 | 10.80 |
ENSRNOT00000060006
|
Ccdc71l
|
coiled-coil domain containing 71-like |
| chr12_-_52072106 | 10.74 |
ENSRNOT00000088306
ENSRNOT00000068621 |
Ddx51
|
DEAD-box helicase 51 |
| chr2_-_30340103 | 10.59 |
ENSRNOT00000024023
ENSRNOT00000067722 |
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr10_+_4536180 | 10.40 |
ENSRNOT00000003309
|
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr5_+_127480485 | 10.35 |
ENSRNOT00000017422
|
Magoh
|
mago homolog, exon junction complex core component |
| chr4_+_6827429 | 10.26 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr11_-_47122095 | 10.18 |
ENSRNOT00000002194
|
Rpl24
|
ribosomal protein L24 |
| chr14_+_44524753 | 10.10 |
ENSRNOT00000076245
|
Rpl9
|
ribosomal protein L9 |
| chr13_+_103300932 | 9.99 |
ENSRNOT00000085214
ENSRNOT00000038264 |
Eprs
|
glutamyl-prolyl-tRNA synthetase |
| chr20_-_4390436 | 9.95 |
ENSRNOT00000000497
ENSRNOT00000077655 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr13_-_109629482 | 9.81 |
ENSRNOT00000072452
|
Flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr10_-_14072230 | 9.68 |
ENSRNOT00000018405
|
Tbl3
|
transducin (beta)-like 3 |
| chr7_-_117773134 | 9.62 |
ENSRNOT00000045135
|
Recql4
|
RecQ like helicase 4 |
| chr12_-_37444282 | 9.59 |
ENSRNOT00000001368
|
Gtf2h3
|
general transcription factor IIH subunit 3 |
| chr12_+_37984790 | 9.32 |
ENSRNOT00000001445
|
Vps37b
|
VPS37B, ESCRT-I subunit |
| chr7_+_27081667 | 9.21 |
ENSRNOT00000066143
|
Nfyb
|
nuclear transcription factor Y subunit beta |
| chr19_-_54722563 | 9.21 |
ENSRNOT00000025784
|
Slc7a5
|
solute carrier family 7 member 5 |
| chr2_-_28370261 | 9.17 |
ENSRNOT00000022259
|
Utp15
|
UTP15, small subunit processome component |
| chr16_-_54899347 | 9.16 |
ENSRNOT00000016321
|
Vps37a
|
VPS37A, ESCRT-I subunit |
| chr10_-_55589978 | 9.11 |
ENSRNOT00000007087
|
LOC100912917
|
phosphoribosylformylglycinamidine synthase-like |
| chr17_+_6840463 | 8.93 |
ENSRNOT00000061233
|
Ubqln1
|
ubiquilin 1 |
| chr8_-_115621394 | 8.91 |
ENSRNOT00000018995
|
Rbm15b
|
RNA binding motif protein 15B |
| chr6_+_26560601 | 8.83 |
ENSRNOT00000077481
|
Gtf3c2
|
general transcription factor IIIC subunit 2 |
| chr3_-_2490392 | 8.83 |
ENSRNOT00000014993
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr9_+_17120759 | 8.79 |
ENSRNOT00000025787
|
Polr1c
|
RNA polymerase I subunit C |
| chr7_-_116872511 | 8.77 |
ENSRNOT00000010120
|
Zc3h3
|
zinc finger CCCH type containing 3 |
| chr1_-_227002139 | 8.75 |
ENSRNOT00000028405
|
Ccdc86
|
coiled-coil domain containing 86 |
| chrX_-_1786978 | 8.68 |
ENSRNOT00000011317
|
Rbm10
|
RNA binding motif protein 10 |
| chr1_-_89124132 | 8.67 |
ENSRNOT00000031044
|
Haus5
|
HAUS augmin-like complex, subunit 5 |
| chr10_-_64398294 | 8.64 |
ENSRNOT00000010386
|
Glod4
|
glyoxalase domain containing 4 |
| chr17_-_13586554 | 8.63 |
ENSRNOT00000041371
|
Secisbp2
|
SECIS binding protein 2 |
| chr4_+_118167294 | 8.41 |
ENSRNOT00000022367
|
LOC687679
|
similar to small nuclear ribonucleoprotein polypeptide G |
| chr20_+_4967194 | 8.37 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr20_-_5865775 | 8.28 |
ENSRNOT00000000612
ENSRNOT00000092641 |
Srpk1
|
SRSF protein kinase 1 |
| chr10_+_103703404 | 8.20 |
ENSRNOT00000086469
|
Rab37
|
RAB37, member RAS oncogene family |
| chrX_-_62698830 | 8.17 |
ENSRNOT00000076359
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr12_+_52072188 | 8.16 |
ENSRNOT00000056725
|
Noc4l
|
nucleolar complex associated 4 homolog |
| chr19_+_37990374 | 8.08 |
ENSRNOT00000026827
|
Dus2
|
dihydrouridine synthase 2 |
| chr10_-_85709568 | 8.06 |
ENSRNOT00000005426
|
Cwc25
|
CWC25 spliceosome-associated protein homolog |
| chr8_+_132204604 | 8.04 |
ENSRNOT00000084725
|
Exosc7
|
exosome component 7 |
| chr5_+_148320438 | 8.01 |
ENSRNOT00000018742
|
Pef1
|
penta-EF hand domain containing 1 |
| chr2_+_187302192 | 8.00 |
ENSRNOT00000015471
|
Isg20l2
|
interferon stimulated exonuclease gene 20-like 2 |
| chr8_+_69127708 | 7.91 |
ENSRNOT00000013490
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr4_-_144620731 | 7.90 |
ENSRNOT00000066468
|
Rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr17_+_43946130 | 7.88 |
ENSRNOT00000023649
|
Abt1
|
activator of basal transcription 1 |
| chr14_+_2789650 | 7.82 |
ENSRNOT00000030479
|
Fam69a
|
family with sequence similarity 69, member A |
| chr7_-_31872423 | 7.80 |
ENSRNOT00000012715
|
Tmpo
|
thymopoietin |
| chr16_+_54899452 | 7.79 |
ENSRNOT00000016882
|
Cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr14_+_80361294 | 7.78 |
ENSRNOT00000011824
|
Trmt44
|
tRNA methyltransferase 44 homolog (S. cerevisiae) |
| chr15_+_47211583 | 7.73 |
ENSRNOT00000016027
|
Pinx1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr1_+_221420271 | 7.67 |
ENSRNOT00000028481
|
LOC687780
|
similar to Finkel-Biskis-Reilly murine sarcoma virusubiquitously expressed |
| chr13_+_99136871 | 7.66 |
ENSRNOT00000078263
ENSRNOT00000004350 |
Sde2
|
SDE2 telomere maintenance homolog |
| chr8_-_72714664 | 7.47 |
ENSRNOT00000024286
|
Rab8b
|
RAB8B, member RAS oncogene family |
| chr15_-_34431325 | 7.37 |
ENSRNOT00000027511
|
Dhrs1
|
dehydrogenase/reductase 1 |
| chr4_-_51946715 | 7.33 |
ENSRNOT00000079130
|
Pot1
|
protection of telomeres 1 |
| chr19_+_49695579 | 7.33 |
ENSRNOT00000016963
|
Gan
|
gigaxonin |
| chr2_+_27365148 | 7.32 |
ENSRNOT00000021549
|
Col4a3bp
|
collagen type IV alpha 3 binding protein |
| chr3_+_7884822 | 7.31 |
ENSRNOT00000019157
|
Med27
|
mediator complex subunit 27 |
| chr6_-_1622196 | 7.29 |
ENSRNOT00000007492
|
Prkd3
|
protein kinase D3 |
| chr16_-_32439421 | 7.20 |
ENSRNOT00000043100
|
Nek1
|
NIMA-related kinase 1 |
| chr2_-_38208719 | 7.19 |
ENSRNOT00000019189
|
Kif2a
|
kinesin family member 2A |
| chr9_+_66045962 | 7.09 |
ENSRNOT00000058491
|
RGD1562399
|
similar to 40S ribosomal protein S2 |
| chr8_-_117811975 | 7.07 |
ENSRNOT00000028055
|
Atrip
|
ATR interacting protein |
| chr1_+_224957517 | 7.06 |
ENSRNOT00000026033
|
Nxf1
|
nuclear RNA export factor 1 |
| chr6_-_96171629 | 7.06 |
ENSRNOT00000010546
ENSRNOT00000078190 |
Trmt5
|
tRNA methyltransferase 5 |
| chr19_+_55300395 | 7.00 |
ENSRNOT00000092169
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr4_+_5841998 | 6.99 |
ENSRNOT00000010025
|
Xrcc2
|
X-ray repair cross complementing 2 |
| chr8_-_118436894 | 6.94 |
ENSRNOT00000073395
|
LOC100911807
|
ATR-interacting protein-like |
| chr20_+_4966817 | 6.91 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_-_57632177 | 6.80 |
ENSRNOT00000080787
ENSRNOT00000092581 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr1_-_64162461 | 6.76 |
ENSRNOT00000091739
|
Prpf31
|
pre-mRNA processing factor 31 |
| chr11_-_64752544 | 6.60 |
ENSRNOT00000048738
|
Tmem39a
|
transmembrane protein 39a |
| chr5_+_117583502 | 6.58 |
ENSRNOT00000010933
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr7_-_60416925 | 6.53 |
ENSRNOT00000008193
|
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr2_-_104133985 | 6.48 |
ENSRNOT00000088167
ENSRNOT00000067725 |
Pde7a
|
phosphodiesterase 7A |
| chr9_+_15393330 | 6.42 |
ENSRNOT00000075630
|
Bysl
|
bystin-like |
| chr4_+_100209951 | 6.36 |
ENSRNOT00000015807
|
LOC691113
|
hypothetical protein LOC691113 |
| chr14_+_44580216 | 6.18 |
ENSRNOT00000088674
ENSRNOT00000003907 |
Rfc1
|
replication factor C subunit 1 |
| chr10_+_105861743 | 6.02 |
ENSRNOT00000064410
|
Mgat5b
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr18_+_1142782 | 5.88 |
ENSRNOT00000046718
|
Thoc1
|
THO complex 1 |
| chr3_-_111560556 | 5.73 |
ENSRNOT00000030532
|
Ltk
|
leukocyte receptor tyrosine kinase |
| chr12_+_36871999 | 5.70 |
ENSRNOT00000001334
ENSRNOT00000084236 |
Ncor2
|
nuclear receptor co-repressor 2 |
| chr17_+_6665659 | 5.62 |
ENSRNOT00000025980
|
Hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr6_-_99400822 | 5.61 |
ENSRNOT00000089594
|
Zbtb25
|
zinc finger and BTB domain containing 25 |
| chr20_-_27308069 | 5.60 |
ENSRNOT00000056047
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr11_+_61748883 | 5.59 |
ENSRNOT00000093552
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr2_+_205553163 | 5.54 |
ENSRNOT00000039572
|
Nras
|
neuroblastoma RAS viral oncogene homolog |
| chr7_+_70328391 | 5.54 |
ENSRNOT00000075544
|
Mettl1
|
methyltransferase like 1 |
| chr19_+_56272162 | 5.51 |
ENSRNOT00000030399
|
Afg3l1
|
AFG3(ATPase family gene 3)-like 1 (S. cerevisiae) |
| chr7_-_126913585 | 5.47 |
ENSRNOT00000036025
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr5_-_136023511 | 5.46 |
ENSRNOT00000081747
|
Rps8
|
ribosomal protein S8 |
| chr8_-_57830504 | 5.43 |
ENSRNOT00000017683
|
Ddx10
|
DEAD-box helicase 10 |
| chr1_-_65551043 | 5.36 |
ENSRNOT00000029996
|
Trim28
|
tripartite motif-containing 28 |
| chr15_+_34520142 | 5.34 |
ENSRNOT00000074659
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr2_-_144007636 | 5.29 |
ENSRNOT00000092028
|
Rfxap
|
regulatory factor X-associated protein |
| chr7_-_137856485 | 5.22 |
ENSRNOT00000007003
|
LOC688906
|
similar to splicing factor, arginine/serine-rich 2, interacting protein |
| chr14_+_2100106 | 5.19 |
ENSRNOT00000000064
|
Gak
|
cyclin G associated kinase |
| chr19_-_55300403 | 5.15 |
ENSRNOT00000018591
|
Rnf166
|
ring finger protein 166 |
| chr3_-_93698121 | 5.14 |
ENSRNOT00000012094
ENSRNOT00000044143 |
Nat10
|
N-acetyltransferase 10 |
| chr6_+_136150785 | 5.09 |
ENSRNOT00000078211
ENSRNOT00000015308 |
LOC103692719
|
tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A |
| chr19_+_14346197 | 5.09 |
ENSRNOT00000043480
|
Gm18025
|
predicted gene, 18025 |
| chr1_+_80028928 | 5.07 |
ENSRNOT00000012082
|
Fbxo46
|
F-box protein 46 |
| chr1_-_199159125 | 4.91 |
ENSRNOT00000025618
|
Bcl7c
|
BCL tumor suppressor 7C |
| chr4_+_99399148 | 4.87 |
ENSRNOT00000088018
|
Rnf103
|
ring finger protein 103 |
| chr10_+_104483042 | 4.85 |
ENSRNOT00000007362
|
Sap30bp
|
SAP30 binding protein |
| chr3_+_138715570 | 4.85 |
ENSRNOT00000064723
|
Sec23b
|
Sec23 homolog B, coat complex II component |
| chr1_-_254671596 | 4.84 |
ENSRNOT00000025450
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr18_+_72550219 | 4.84 |
ENSRNOT00000046847
|
Smad2
|
SMAD family member 2 |
| chr10_+_91200874 | 4.78 |
ENSRNOT00000004272
|
Hexim1
|
hexamethylene bis-acetamide inducible 1 |
| chr2_+_264864351 | 4.77 |
ENSRNOT00000015434
|
Lrrc40
|
leucine rich repeat containing 40 |
| chr12_+_17277635 | 4.75 |
ENSRNOT00000001734
|
LOC498154
|
hypothetical protein LOC498154 |
| chr12_-_46989876 | 4.73 |
ENSRNOT00000079043
ENSRNOT00000001534 |
NEWGENE_1586233
|
TP53 regulated inhibitor of apoptosis 1 |
| chr7_+_99677290 | 4.72 |
ENSRNOT00000086039
ENSRNOT00000005376 |
Nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr2_-_144217600 | 4.71 |
ENSRNOT00000000101
|
Rfxapl1
|
regulatory factor X-associated protein-like 1 |
| chr14_+_100415668 | 4.69 |
ENSRNOT00000008057
|
C1d
|
C1D nuclear receptor co-repressor |
| chr1_-_89042176 | 4.63 |
ENSRNOT00000080842
|
Kmt2b
|
lysine methyltransferase 2B |
| chr2_-_264864265 | 4.63 |
ENSRNOT00000044236
|
Srsf11
|
serine and arginine rich splicing factor 11 |
| chr20_-_10257044 | 4.59 |
ENSRNOT00000068289
|
Wdr4
|
WD repeat domain 4 |
| chr12_+_24669449 | 4.53 |
ENSRNOT00000044449
|
Wbscr22
|
Williams Beuren syndrome chromosome region 22 |
| chr12_-_47031498 | 4.52 |
ENSRNOT00000075320
|
Triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr19_-_37990353 | 4.50 |
ENSRNOT00000026817
|
Ddx28
|
DEAD-box helicase 28 |
| chr20_+_8325041 | 4.47 |
ENSRNOT00000000640
|
Cmtr1
|
cap methyltransferase 1 |
| chr1_-_242861767 | 4.46 |
ENSRNOT00000021185
|
Cbwd1
|
COBW domain containing 1 |
| chr17_-_10527839 | 4.43 |
ENSRNOT00000064477
ENSRNOT00000091837 |
Faf2
|
Fas associated factor family member 2 |
| chr6_+_135610743 | 4.39 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr1_-_63293635 | 4.38 |
ENSRNOT00000090337
|
Zik1
|
zinc finger protein interacting with K protein 1 |
| chr7_-_99677268 | 4.27 |
ENSRNOT00000013315
|
RGD1564420
|
similar to Hypothetical protein MGC31278 |
| chr19_+_22569999 | 4.24 |
ENSRNOT00000022573
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr9_-_17120677 | 4.12 |
ENSRNOT00000025769
|
Yipf3
|
Yip1 domain family, member 3 |
| chr18_+_72550454 | 4.11 |
ENSRNOT00000092173
|
Smad2
|
SMAD family member 2 |
| chr2_-_27364906 | 4.10 |
ENSRNOT00000078639
|
Polk
|
DNA polymerase kappa |
| chr6_-_109692218 | 4.07 |
ENSRNOT00000012327
|
RGD1310769
|
similar to HSPC288 |
| chrX_-_45522665 | 3.92 |
ENSRNOT00000030771
|
RGD1562200
|
similar to GS2 gene |
| chr3_-_110964452 | 3.90 |
ENSRNOT00000016356
|
Rmdn3
|
regulator of microtubule dynamics 3 |
| chr19_+_31867981 | 3.86 |
ENSRNOT00000024753
|
Abce1
|
ATP binding cassette subfamily E member 1 |
| chr9_+_82411013 | 3.82 |
ENSRNOT00000026194
|
Stk16
|
serine/threonine kinase 16 |
| chr10_+_38692211 | 3.71 |
ENSRNOT00000009440
|
Aff4
|
AF4/FMR2 family, member 4 |
| chr20_-_3299580 | 3.56 |
ENSRNOT00000050373
|
Gnl1
|
G protein nucleolar 1 |
| chr12_-_2170504 | 3.50 |
ENSRNOT00000001309
|
Xab2
|
XPA binding protein 2 |
| chr19_-_31867850 | 3.49 |
ENSRNOT00000024689
|
Anapc10
|
anaphase promoting complex subunit 10 |
| chr1_+_56982821 | 3.46 |
ENSRNOT00000059845
|
Ermard
|
ER membrane-associated RNA degradation |
| chr1_-_190370499 | 3.44 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr3_+_58443101 | 3.41 |
ENSRNOT00000002075
|
Itga6
|
integrin subunit alpha 6 |
| chr2_+_196277978 | 3.41 |
ENSRNOT00000028625
|
Vps72
|
vacuolar protein sorting 72 homolog |
| chr15_+_58170967 | 3.40 |
ENSRNOT00000001366
|
Nufip1
|
NUFIP1, FMR1 interacting protein 1 |
| chr13_+_25778317 | 3.39 |
ENSRNOT00000021129
|
Tnfrsf11a
|
TNF receptor superfamily member 11A |
| chr20_+_28027054 | 3.36 |
ENSRNOT00000071386
ENSRNOT00000001044 |
Ranbp2
|
RAN binding protein 2 |
| chr8_+_114866768 | 3.32 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr2_-_211638788 | 3.27 |
ENSRNOT00000027666
|
Stxbp3
|
syntaxin binding protein 3 |
| chr11_+_87522971 | 3.25 |
ENSRNOT00000043545
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr20_-_3299420 | 3.14 |
ENSRNOT00000090999
|
Gnl1
|
G protein nucleolar 1 |
| chr20_+_3299730 | 3.11 |
ENSRNOT00000078538
|
Prr3
|
proline rich 3 |
| chr11_-_86890390 | 3.04 |
ENSRNOT00000046183
|
Trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr19_+_43251539 | 3.02 |
ENSRNOT00000024793
|
Ddx19a
|
DEAD-box helicase 19A |
| chr1_+_1897338 | 2.99 |
ENSRNOT00000021437
|
Ppil4
|
peptidylprolyl isomerase like 4 |
| chrX_-_155862363 | 2.97 |
ENSRNOT00000089847
|
Dkc1
|
dyskerin pseudouridine synthase 1 |
| chr20_+_6205903 | 2.94 |
ENSRNOT00000092333
ENSRNOT00000092655 |
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr6_-_43493816 | 2.93 |
ENSRNOT00000077786
|
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr3_+_112519808 | 2.90 |
ENSRNOT00000014129
|
Rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr5_+_144701949 | 2.87 |
ENSRNOT00000015618
|
Psmb2
|
proteasome subunit beta 2 |
| chr3_-_13896838 | 2.87 |
ENSRNOT00000025399
|
Fbxw2
|
F-box and WD repeat domain containing 2 |
| chr10_+_55924938 | 2.84 |
ENSRNOT00000087003
ENSRNOT00000057079 |
Trappc1
|
trafficking protein particle complex 1 |
| chr17_-_9695292 | 2.83 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr1_-_68269117 | 2.80 |
ENSRNOT00000072367
|
LOC100911224
|
zinc finger protein interacting with ribonucleoprotein K-like |
| chr1_-_254671778 | 2.76 |
ENSRNOT00000025493
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr7_+_117773230 | 2.76 |
ENSRNOT00000077562
|
Lrrc14
|
leucine rich repeat containing 14 |
| chr10_+_64398339 | 2.73 |
ENSRNOT00000056278
|
Mrm3
|
mitochondrial rRNA methyltransferase 3 |
| chr3_+_111097326 | 2.69 |
ENSRNOT00000018717
|
Vps18
|
VPS18 CORVET/HOPS core subunit |
| chr5_+_57291156 | 2.66 |
ENSRNOT00000060692
ENSRNOT00000060691 |
Nfx1
|
nuclear transcription factor, X-box binding 1 |
| chr1_-_214163808 | 2.65 |
ENSRNOT00000082284
|
Rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr15_+_24056383 | 2.59 |
ENSRNOT00000015120
|
Socs4
|
suppressor of cytokine signaling 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Creb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.7 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 6.1 | 18.4 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 5.6 | 16.9 | GO:0014016 | neuroblast differentiation(GO:0014016) spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) medulla oblongata development(GO:0021550) musculoskeletal movement, spinal reflex action(GO:0050883) olfactory pit development(GO:0060166) |
| 3.3 | 10.0 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 3.3 | 9.8 | GO:0097037 | heme export(GO:0097037) |
| 3.2 | 15.8 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 3.1 | 9.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 3.0 | 8.9 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 2.9 | 14.6 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) lens fiber cell apoptotic process(GO:1990086) |
| 2.9 | 8.6 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 2.9 | 31.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 2.8 | 22.0 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 2.7 | 8.0 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 2.6 | 18.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 2.6 | 7.7 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 2.5 | 10.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 2.5 | 10.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 2.4 | 17.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 2.4 | 7.3 | GO:0060383 | positive regulation of DNA strand elongation(GO:0060383) |
| 2.4 | 12.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 2.4 | 7.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 2.3 | 7.0 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 2.3 | 29.7 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 2.3 | 11.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 2.2 | 8.9 | GO:1900224 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 2.2 | 8.8 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 2.0 | 8.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 2.0 | 11.9 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 2.0 | 7.9 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 1.9 | 9.6 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 1.9 | 7.6 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 1.9 | 13.3 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 1.9 | 5.6 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 1.8 | 5.5 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.8 | 5.4 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 1.7 | 5.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 1.6 | 11.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 1.6 | 17.5 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 1.5 | 4.5 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 1.5 | 7.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 1.4 | 4.2 | GO:0042196 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 1.4 | 5.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 1.4 | 4.1 | GO:0016129 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 1.4 | 12.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 1.4 | 6.8 | GO:0048254 | snoRNA localization(GO:0048254) |
| 1.3 | 8.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 1.3 | 5.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 1.3 | 10.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 1.3 | 12.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 1.3 | 3.8 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 1.2 | 7.5 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 1.2 | 9.9 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 1.2 | 7.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 1.2 | 7.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) strand invasion(GO:0042148) |
| 1.1 | 3.4 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 1.1 | 4.5 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 1.0 | 6.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 1.0 | 2.0 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 1.0 | 3.0 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 1.0 | 2.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 1.0 | 5.9 | GO:0045829 | negative regulation of isotype switching(GO:0045829) regulation of isotype switching to IgA isotypes(GO:0048296) negative regulation of DNA damage checkpoint(GO:2000002) |
| 1.0 | 3.9 | GO:0060701 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.9 | 4.7 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.9 | 9.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.9 | 5.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.8 | 8.7 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.8 | 29.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.7 | 8.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.7 | 5.6 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.7 | 14.6 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.7 | 3.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.7 | 12.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.7 | 2.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.7 | 2.7 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.7 | 7.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.6 | 7.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.6 | 5.7 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.6 | 7.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.6 | 1.7 | GO:1902953 | endoplasmic reticulum membrane organization(GO:0090158) positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.5 | 1.6 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.5 | 2.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.5 | 4.5 | GO:2001138 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.5 | 5.5 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.5 | 4.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.5 | 8.0 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.5 | 3.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.4 | 5.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.4 | 19.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.4 | 7.9 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.4 | 16.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.4 | 15.0 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.4 | 23.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.4 | 6.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.4 | 3.4 | GO:0010668 | ectodermal cell differentiation(GO:0010668) nail development(GO:0035878) |
| 0.4 | 3.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.3 | 3.5 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.3 | 3.8 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 1.3 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.3 | 5.5 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.3 | 4.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 5.7 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.3 | 2.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.3 | 9.9 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.3 | 3.3 | GO:0022615 | protein to membrane docking(GO:0022615) negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.3 | 0.8 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.3 | 9.1 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.3 | 6.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.3 | 4.8 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.2 | 8.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 4.7 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.2 | 5.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 4.8 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.2 | 2.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.2 | 9.2 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.2 | 14.0 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.2 | 2.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 1.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.2 | 12.1 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 3.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 1.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 6.4 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.2 | 15.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 0.8 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 2.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.4 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 5.1 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.1 | 3.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 8.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 1.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 3.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.7 | GO:0032438 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.1 | 5.4 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.1 | 4.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.3 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 3.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 2.4 | GO:0048536 | spleen development(GO:0048536) |
| 0.1 | 8.0 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.1 | 7.3 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.1 | 0.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 2.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 10.6 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 2.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 2.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.6 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 15.0 | GO:0006974 | cellular response to DNA damage stimulus(GO:0006974) |
| 0.0 | 0.4 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 1.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.2 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 7.0 | GO:0072659 | protein localization to plasma membrane(GO:0072659) |
| 0.0 | 1.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.7 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 3.2 | GO:0071560 | cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 19.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 3.2 | 9.6 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 3.1 | 15.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 3.0 | 15.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 2.4 | 14.6 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 2.2 | 8.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 2.2 | 19.4 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 2.1 | 18.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 1.9 | 5.6 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 1.8 | 5.5 | GO:0005745 | m-AAA complex(GO:0005745) |
| 1.7 | 17.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 1.7 | 16.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 1.6 | 8.2 | GO:0030689 | Noc complex(GO:0030689) |
| 1.6 | 9.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 1.5 | 18.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.4 | 7.0 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.3 | 9.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 1.3 | 16.5 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 1.3 | 15.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 1.2 | 10.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.2 | 16.6 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 1.0 | 6.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 1.0 | 8.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.9 | 6.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.9 | 12.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.9 | 12.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.9 | 18.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.9 | 8.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.8 | 3.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.8 | 3.3 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.7 | 7.3 | GO:0070187 | telosome(GO:0070187) |
| 0.7 | 2.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.6 | 12.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.6 | 7.5 | GO:0051286 | cell tip(GO:0051286) |
| 0.6 | 27.4 | GO:0000791 | euchromatin(GO:0000791) |
| 0.6 | 3.0 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.6 | 4.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.6 | 5.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.6 | 8.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.5 | 4.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.5 | 22.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.5 | 44.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 7.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.4 | 0.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.4 | 3.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.4 | 10.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.4 | 4.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.4 | 2.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 2.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 15.8 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.3 | 8.0 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.3 | 3.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.3 | 2.7 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.3 | 17.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.3 | 7.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 5.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 12.0 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.2 | 2.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 5.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.2 | 4.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 7.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 4.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 3.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 8.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 3.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 15.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 7.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 5.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 6.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 2.0 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 7.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 10.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 27.3 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 9.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 2.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 6.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 4.6 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 22.3 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 7.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 9.1 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 28.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 9.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 6.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 3.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 7.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 10.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 7.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 3.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.6 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 12.5 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 7.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 17.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 5.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 6.6 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 9.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 5.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 5.5 | 22.0 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 4.2 | 25.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 4.0 | 7.9 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 3.6 | 14.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 3.3 | 10.0 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 2.6 | 10.6 | GO:0001156 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIC-class transcription factor binding(GO:0001156) |
| 2.2 | 8.9 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 2.2 | 13.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 2.2 | 15.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 2.0 | 18.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 2.0 | 17.6 | GO:0043495 | protein anchor(GO:0043495) |
| 1.9 | 7.8 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 1.8 | 5.4 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 1.8 | 7.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 1.7 | 5.1 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 1.7 | 6.8 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 1.7 | 10.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 1.7 | 16.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 1.5 | 4.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.5 | 6.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 1.5 | 4.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 1.4 | 8.6 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 1.4 | 9.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 1.4 | 5.6 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 1.3 | 8.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 1.3 | 3.9 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 1.2 | 7.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 1.2 | 8.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 1.2 | 7.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 1.1 | 5.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 1.1 | 9.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.1 | 6.5 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 1.0 | 25.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 1.0 | 10.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.9 | 7.5 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.8 | 15.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.8 | 3.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.7 | 4.4 | GO:0035473 | lipase binding(GO:0035473) |
| 0.7 | 5.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.7 | 3.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.7 | 2.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.7 | 12.8 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.6 | 2.5 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.6 | 6.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.6 | 23.8 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.5 | 3.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.5 | 7.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.5 | 8.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.5 | 16.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.5 | 10.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 25.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.4 | 11.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.4 | 12.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.4 | 7.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.4 | 7.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.4 | 6.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.4 | 10.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.4 | 33.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.3 | 8.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 13.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.3 | 3.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.3 | 5.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 8.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 16.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.3 | 4.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.3 | 12.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.3 | 6.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 18.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 9.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 5.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 1.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 4.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 4.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 11.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 12.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.2 | 4.8 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.2 | 9.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.2 | 3.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 1.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 2.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 5.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 8.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 2.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 10.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 3.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 18.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 135.4 | GO:0003723 | RNA binding(GO:0003723) |
| 0.1 | 5.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 1.6 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 1.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 3.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 6.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 7.8 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 2.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 6.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 3.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 4.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 10.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 15.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 2.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 2.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 5.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 25.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 1.0 | 7.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.9 | 14.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.8 | 34.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.5 | 5.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 22.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 18.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.3 | 15.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 3.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 18.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.3 | 13.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 12.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 12.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 7.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 6.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 4.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 10.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 7.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 11.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 0.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.8 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 34.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 2.1 | 18.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 1.4 | 39.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 1.1 | 10.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 1.0 | 24.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 1.0 | 10.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 1.0 | 15.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 1.0 | 30.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.9 | 7.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.9 | 7.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.8 | 34.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.8 | 17.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.7 | 7.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.7 | 14.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.6 | 8.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.6 | 22.9 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.6 | 5.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.5 | 7.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.5 | 49.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.4 | 5.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.4 | 14.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.4 | 4.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.4 | 6.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.4 | 10.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 7.8 | REACTOME DEADENYLATION DEPENDENT MRNA DECAY | Genes involved in Deadenylation-dependent mRNA decay |
| 0.3 | 9.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 4.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.3 | 2.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 6.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 9.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 3.5 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.2 | 1.9 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.2 | 7.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 2.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 8.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.0 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.1 | 17.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 2.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 7.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 3.3 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 1.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 16.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 6.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 2.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 3.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |