Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
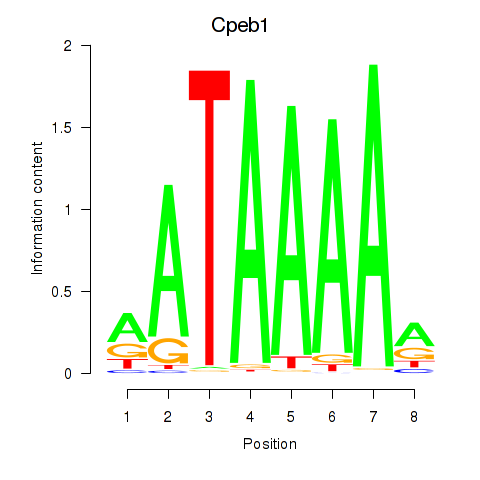
Results for Cpeb1
Z-value: 1.13
Transcription factors associated with Cpeb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cpeb1
|
ENSRNOG00000019161 | cytoplasmic polyadenylation element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cpeb1 | rn6_v1_chr1_-_143256817_143256817 | 0.60 | 7.0e-33 | Click! |
Activity profile of Cpeb1 motif
Sorted Z-values of Cpeb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_93949187 | 67.33 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr3_+_98297554 | 54.08 |
ENSRNOT00000006524
|
Kcna4
|
potassium voltage-gated channel subfamily A member 4 |
| chr20_-_54269525 | 52.73 |
ENSRNOT00000037559
|
Grik2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr2_+_27905535 | 48.06 |
ENSRNOT00000022120
|
Fam169a
|
family with sequence similarity 169, member A |
| chr3_+_129599353 | 39.39 |
ENSRNOT00000008734
|
Snap25
|
synaptosomal-associated protein 25 |
| chr7_-_29701586 | 39.04 |
ENSRNOT00000009084
ENSRNOT00000089269 |
Ano4
|
anoctamin 4 |
| chr10_-_27366665 | 38.14 |
ENSRNOT00000004725
|
Gabra1
|
gamma-aminobutyric acid type A receptor alpha1 subunit |
| chr6_+_113898420 | 37.08 |
ENSRNOT00000064872
|
Nrxn3
|
neurexin 3 |
| chr6_-_136550371 | 35.54 |
ENSRNOT00000065971
|
Rd3l
|
retinal degeneration 3-like |
| chr2_-_172361779 | 35.44 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr4_-_108717309 | 32.38 |
ENSRNOT00000085062
|
AABR07061178.1
|
|
| chr9_+_73334618 | 31.86 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr9_-_32019205 | 31.59 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr7_+_64768742 | 30.67 |
ENSRNOT00000005545
|
Grip1
|
glutamate receptor interacting protein 1 |
| chrX_+_122808605 | 29.07 |
ENSRNOT00000017567
|
Zcchc12
|
zinc finger CCHC-type containing 12 |
| chr2_+_136993208 | 28.99 |
ENSRNOT00000040187
ENSRNOT00000066542 |
Pcdh10
|
protocadherin 10 |
| chr13_-_88061108 | 28.48 |
ENSRNOT00000003774
|
Rgs4
|
regulator of G-protein signaling 4 |
| chr2_+_184230459 | 28.43 |
ENSRNOT00000074187
|
AABR07012054.1
|
|
| chr17_-_18590536 | 28.33 |
ENSRNOT00000078992
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr10_-_18579632 | 26.16 |
ENSRNOT00000040737
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr3_+_48096954 | 25.87 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr18_-_37245809 | 24.75 |
ENSRNOT00000079585
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr5_-_85123829 | 24.05 |
ENSRNOT00000007578
|
Brinp1
|
BMP/retinoic acid inducible neural specific 1 |
| chr8_+_23398030 | 23.38 |
ENSRNOT00000031893
|
RGD1561444
|
similar to RIKEN cDNA 9530077C05 |
| chr14_-_85191557 | 23.30 |
ENSRNOT00000011604
|
Nefh
|
neurofilament heavy |
| chr5_-_65073012 | 23.08 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr15_+_17834635 | 23.07 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr13_+_57243877 | 23.05 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr11_-_35098883 | 22.52 |
ENSRNOT00000079955
|
Kcnj6
|
potassium voltage-gated channel subfamily J member 6 |
| chr5_-_12563429 | 22.05 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr20_-_5064469 | 21.18 |
ENSRNOT00000001120
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr9_+_73529612 | 21.00 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr18_-_18079560 | 20.20 |
ENSRNOT00000072093
|
AABR07031533.1
|
|
| chr2_+_187322416 | 19.72 |
ENSRNOT00000025183
|
Crabp2
|
cellular retinoic acid binding protein 2 |
| chr2_+_263212051 | 19.58 |
ENSRNOT00000089396
|
Negr1
|
neuronal growth regulator 1 |
| chr7_-_136853154 | 19.54 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chr2_-_98610368 | 19.07 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chrX_-_152642531 | 18.51 |
ENSRNOT00000085037
|
Gabra3
|
gamma-aminobutyric acid type A receptor alpha3 subunit |
| chr6_-_8344574 | 17.49 |
ENSRNOT00000009660
|
Prepl
|
prolyl endopeptidase-like |
| chr4_-_482645 | 17.40 |
ENSRNOT00000062073
ENSRNOT00000071713 |
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr13_-_89565813 | 17.14 |
ENSRNOT00000004347
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr10_+_80790168 | 17.07 |
ENSRNOT00000073315
ENSRNOT00000075163 |
Car10
|
carbonic anhydrase 10 |
| chr13_-_82753438 | 16.97 |
ENSRNOT00000075948
|
Atp1b1
|
ATPase Na+/K+ transporting subunit beta 1 |
| chr19_-_11669578 | 16.88 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr5_-_168734296 | 16.31 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr3_-_51612397 | 16.16 |
ENSRNOT00000081401
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr1_-_174411141 | 16.15 |
ENSRNOT00000065288
|
Nrip3
|
nuclear receptor interacting protein 3 |
| chr20_-_47910375 | 16.08 |
ENSRNOT00000000348
|
Sobp
|
sine oculis binding protein homolog |
| chr6_+_58468155 | 15.99 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chrX_-_142164220 | 15.90 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr3_+_100786862 | 15.32 |
ENSRNOT00000080190
|
Bdnf
|
brain-derived neurotrophic factor |
| chr2_-_149444548 | 15.02 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr2_+_207930796 | 14.61 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr7_-_70476340 | 14.61 |
ENSRNOT00000006800
|
Arhgef25
|
Rho guanine nucleotide exchange factor 25 |
| chr7_+_83113672 | 14.43 |
ENSRNOT00000006783
|
Trhr
|
thyrotropin releasing hormone receptor |
| chr7_-_119352605 | 14.30 |
ENSRNOT00000008414
|
Cacng2
|
calcium voltage-gated channel auxiliary subunit gamma 2 |
| chr6_-_23291568 | 13.95 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr20_+_5049496 | 13.71 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr18_+_13386133 | 13.66 |
ENSRNOT00000020661
|
Asxl3
|
additional sex combs like 3, transcriptional regulator |
| chr5_+_152680407 | 13.43 |
ENSRNOT00000076864
|
Stmn1
|
stathmin 1 |
| chr15_-_80713153 | 13.35 |
ENSRNOT00000063800
|
Klhl1
|
kelch-like family member 1 |
| chr9_+_100489852 | 13.18 |
ENSRNOT00000022854
|
Ppp1r7
|
protein phosphatase 1, regulatory subunit 7 |
| chr2_-_2814690 | 12.67 |
ENSRNOT00000029265
|
Gpr150
|
G protein-coupled receptor 150 |
| chr9_-_79898912 | 12.50 |
ENSRNOT00000022076
|
March4
|
membrane associated ring-CH-type finger 4 |
| chr15_-_43542939 | 12.30 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr9_-_121713091 | 12.29 |
ENSRNOT00000073432
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr4_+_3959640 | 12.25 |
ENSRNOT00000009439
|
Paxip1
|
PAX interacting protein 1 |
| chr6_-_8344897 | 12.15 |
ENSRNOT00000082353
|
Prepl
|
prolyl endopeptidase-like |
| chr5_+_2278357 | 11.89 |
ENSRNOT00000066590
|
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr20_+_3422461 | 11.69 |
ENSRNOT00000084917
ENSRNOT00000079854 |
Tubb5
|
tubulin, beta 5 class I |
| chr7_-_116255167 | 11.59 |
ENSRNOT00000038109
ENSRNOT00000041774 |
Cyp11b2
|
cytochrome P450, family 11, subfamily b, polypeptide 2 |
| chr7_+_44009069 | 11.59 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr10_+_77537340 | 11.40 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chr7_-_116161708 | 10.75 |
ENSRNOT00000068707
|
Cyp11b2
|
cytochrome P450, family 11, subfamily b, polypeptide 2 |
| chr2_+_22910236 | 10.72 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr19_+_20147037 | 10.41 |
ENSRNOT00000020028
|
Zfp423
|
zinc finger protein 423 |
| chrX_+_51286737 | 10.34 |
ENSRNOT00000035692
|
Dmd
|
dystrophin |
| chr1_+_142679345 | 10.20 |
ENSRNOT00000034267
|
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr6_+_8669722 | 10.17 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr2_-_186232292 | 10.04 |
ENSRNOT00000087088
|
Dclk2
|
doublecortin-like kinase 2 |
| chr2_-_154418629 | 10.01 |
ENSRNOT00000076274
ENSRNOT00000076165 |
Plch1
|
phospholipase C, eta 1 |
| chr2_+_239415046 | 9.93 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr8_-_44327551 | 9.64 |
ENSRNOT00000083939
|
Gramd1b
|
GRAM domain containing 1B |
| chr1_+_64928503 | 9.57 |
ENSRNOT00000086274
|
Vom2r80
|
vomeronasal 2 receptor, 80 |
| chr9_+_88494676 | 9.55 |
ENSRNOT00000089451
|
LOC102556337
|
mitochondrial fission factor-like |
| chr3_+_47677720 | 9.55 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr10_-_64657089 | 9.37 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr20_+_25990304 | 9.33 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr2_-_154418920 | 9.23 |
ENSRNOT00000076326
|
Plch1
|
phospholipase C, eta 1 |
| chrX_-_30831483 | 9.22 |
ENSRNOT00000004443
|
Glra2
|
glycine receptor, alpha 2 |
| chr8_+_98745310 | 9.08 |
ENSRNOT00000019938
|
Zic4
|
Zic family member 4 |
| chr8_+_45797315 | 8.47 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr11_+_31389514 | 8.45 |
ENSRNOT00000000325
|
Olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr6_+_94835845 | 8.35 |
ENSRNOT00000006321
|
Jkamp
|
JNK1/MAPK8-associated membrane protein |
| chr9_-_88534710 | 8.32 |
ENSRNOT00000020880
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr18_-_16542165 | 8.16 |
ENSRNOT00000079381
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr8_-_60570058 | 8.16 |
ENSRNOT00000009169
|
Scaper
|
S-phase cyclin A-associated protein in the ER |
| chr4_-_79989572 | 8.11 |
ENSRNOT00000013264
ENSRNOT00000086453 |
Dfna5
|
deafness, autosomal dominant 5 (human) |
| chr4_-_29778039 | 8.10 |
ENSRNOT00000074177
|
Sgce
|
sarcoglycan, epsilon |
| chr18_+_65285318 | 8.06 |
ENSRNOT00000020431
|
Tcf4
|
transcription factor 4 |
| chr14_+_104250617 | 8.05 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr3_-_61488696 | 7.94 |
ENSRNOT00000083045
|
Lnpk
|
lunapark, ER junction formation factor |
| chr1_-_250626844 | 7.90 |
ENSRNOT00000077135
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr5_+_58667885 | 7.81 |
ENSRNOT00000064755
|
Unc13b
|
unc-13 homolog B |
| chr11_-_782954 | 7.76 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr3_+_95715193 | 7.73 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr20_-_10013190 | 7.60 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chr5_+_139394794 | 7.49 |
ENSRNOT00000045954
|
Scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr5_+_4373626 | 7.45 |
ENSRNOT00000064946
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr7_-_94563001 | 7.43 |
ENSRNOT00000051139
ENSRNOT00000005561 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr4_-_23718047 | 7.36 |
ENSRNOT00000011359
|
RGD1564345
|
similar to RIKEN cDNA 4921511H03 |
| chr9_-_19613360 | 7.27 |
ENSRNOT00000029593
|
Rcan2
|
regulator of calcineurin 2 |
| chr18_+_29993361 | 7.23 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chrX_+_984798 | 7.20 |
ENSRNOT00000073016
|
Zfp182
|
zinc finger protein 182 |
| chr18_+_59748444 | 7.19 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr8_-_67040005 | 7.11 |
ENSRNOT00000038641
|
Glce
|
glucuronic acid epimerase |
| chr7_+_64769089 | 7.11 |
ENSRNOT00000088861
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr2_-_192027225 | 7.07 |
ENSRNOT00000016430
|
Prr9
|
proline rich 9 |
| chr15_-_88036354 | 6.98 |
ENSRNOT00000014747
|
Ednrb
|
endothelin receptor type B |
| chr7_-_33916338 | 6.95 |
ENSRNOT00000005492
|
Cfap54
|
cilia and flagella associated protein 54 |
| chr17_+_44528125 | 6.95 |
ENSRNOT00000084538
|
LOC680322
|
similar to Histone H2A type 1 |
| chr2_+_216863428 | 6.83 |
ENSRNOT00000068413
|
Col11a1
|
collagen type XI alpha 1 chain |
| chr10_-_73629581 | 6.78 |
ENSRNOT00000091172
|
Brip1
|
BRCA1 interacting protein C-terminal helicase 1 |
| chr1_-_246110218 | 6.77 |
ENSRNOT00000077544
|
Rfx3
|
regulatory factor X3 |
| chr8_-_47404010 | 6.76 |
ENSRNOT00000038647
|
Tmem136
|
transmembrane protein 136 |
| chr2_-_234296145 | 6.76 |
ENSRNOT00000014155
|
Elovl6
|
ELOVL fatty acid elongase 6 |
| chr2_+_260249566 | 6.75 |
ENSRNOT00000089358
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr16_-_9658484 | 6.68 |
ENSRNOT00000065216
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr15_-_48284548 | 6.54 |
ENSRNOT00000038336
|
Hmbox1
|
homeobox containing 1 |
| chr3_-_157049666 | 6.50 |
ENSRNOT00000089958
|
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr16_-_32421005 | 6.39 |
ENSRNOT00000082712
|
Nek1
|
NIMA-related kinase 1 |
| chr10_-_87136026 | 6.37 |
ENSRNOT00000014230
ENSRNOT00000083233 |
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr1_+_30681681 | 6.28 |
ENSRNOT00000015395
|
Rspo3
|
R-spondin 3 |
| chr5_-_155204456 | 6.28 |
ENSRNOT00000089574
|
Ephb2
|
Eph receptor B2 |
| chr12_+_18931477 | 6.26 |
ENSRNOT00000077087
|
Spry3
|
sprouty RTK signaling antagonist 3 |
| chr7_+_144865608 | 6.18 |
ENSRNOT00000091596
ENSRNOT00000055285 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr6_+_6946695 | 6.09 |
ENSRNOT00000061921
|
Mta3
|
metastasis associated 1 family, member 3 |
| chr15_-_2966576 | 6.05 |
ENSRNOT00000070893
ENSRNOT00000017383 |
Kat6b
|
lysine acetyltransferase 6B |
| chr17_-_22143324 | 6.04 |
ENSRNOT00000019361
|
Edn1
|
endothelin 1 |
| chr16_-_20939545 | 5.98 |
ENSRNOT00000027457
|
Sugp2
|
SURP and G patch domain containing 2 |
| chr17_-_21484456 | 5.97 |
ENSRNOT00000038180
|
Sycp2l
|
synaptonemal complex protein 2-like |
| chr18_+_30820321 | 5.71 |
ENSRNOT00000060472
|
Pcdhga3
|
protocadherin gamma subfamily A, 3 |
| chr5_+_58855773 | 5.64 |
ENSRNOT00000072869
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr14_+_69800156 | 5.60 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr2_+_234375315 | 5.59 |
ENSRNOT00000071270
|
LOC102549542
|
elongation of very long chain fatty acids protein 6-like |
| chr5_-_6186329 | 5.59 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr5_-_7874909 | 5.57 |
ENSRNOT00000064774
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr10_+_76320041 | 5.56 |
ENSRNOT00000037693
|
Coil
|
coilin |
| chr3_+_137154086 | 5.55 |
ENSRNOT00000034252
|
Otor
|
otoraplin |
| chrX_+_156655960 | 5.52 |
ENSRNOT00000085723
|
Mecp2
|
methyl CpG binding protein 2 |
| chr10_+_90230441 | 5.44 |
ENSRNOT00000082722
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr1_+_221596148 | 5.43 |
ENSRNOT00000028536
|
Gpha2
|
glycoprotein hormone alpha 2 |
| chr5_+_164796185 | 5.39 |
ENSRNOT00000010779
|
Nppb
|
natriuretic peptide B |
| chr8_-_18762922 | 5.18 |
ENSRNOT00000008423
|
Olr1122
|
olfactory receptor 1122 |
| chr15_+_37171052 | 5.11 |
ENSRNOT00000011684
|
Zmym2
|
zinc finger MYM-type containing 2 |
| chr18_-_26211445 | 5.03 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chrX_-_9999401 | 5.03 |
ENSRNOT00000060992
|
Gpr82
|
G protein-coupled receptor 82 |
| chr18_+_40853988 | 5.02 |
ENSRNOT00000091759
|
AABR07031963.1
|
uncharacterized protein LOC317165 |
| chr9_+_2190915 | 4.94 |
ENSRNOT00000077417
|
Satb1
|
SATB homeobox 1 |
| chr9_-_44419998 | 4.89 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr14_-_19159923 | 4.86 |
ENSRNOT00000003879
|
Afp
|
alpha-fetoprotein |
| chr3_-_157099306 | 4.83 |
ENSRNOT00000022861
|
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr4_+_118207862 | 4.80 |
ENSRNOT00000085787
ENSRNOT00000023103 |
Tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr1_-_20962526 | 4.73 |
ENSRNOT00000061332
ENSRNOT00000017322 ENSRNOT00000017412 ENSRNOT00000079688 ENSRNOT00000017417 |
Epb41l2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr10_+_90230711 | 4.71 |
ENSRNOT00000055185
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr9_+_51298426 | 4.68 |
ENSRNOT00000065212
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr8_+_5522739 | 4.59 |
ENSRNOT00000011507
|
Mmp13
|
matrix metallopeptidase 13 |
| chr4_-_2201749 | 4.43 |
ENSRNOT00000089327
|
Lmbr1
|
limb development membrane protein 1 |
| chr1_-_255815733 | 4.42 |
ENSRNOT00000047387
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr10_+_35343189 | 4.34 |
ENSRNOT00000083688
|
Mapk9
|
mitogen-activated protein kinase 9 |
| chr3_+_100787449 | 4.28 |
ENSRNOT00000078543
|
Bdnf
|
brain-derived neurotrophic factor |
| chr11_+_73022063 | 4.22 |
ENSRNOT00000002361
|
Ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr8_-_6235967 | 4.05 |
ENSRNOT00000068290
|
LOC654482
|
hypothetical protein LOC654482 |
| chr2_+_149843282 | 4.03 |
ENSRNOT00000074805
|
RGD1561998
|
similar to hypothetical protein C130079G13 |
| chr4_+_2055615 | 3.97 |
ENSRNOT00000007646
|
Rnf32
|
ring finger protein 32 |
| chr11_-_81379640 | 3.94 |
ENSRNOT00000002484
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr3_+_150135821 | 3.93 |
ENSRNOT00000047286
|
Zfp341
|
zinc finger protein 341 |
| chr4_-_77510202 | 3.93 |
ENSRNOT00000038550
|
Zfp786
|
zinc finger protein 786 |
| chr2_+_22909569 | 3.91 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr19_-_29016521 | 3.89 |
ENSRNOT00000049860
|
LOC685160
|
similar to spermatogenesis associated glutamate (E)-rich protein 4d |
| chr17_-_9458288 | 3.84 |
ENSRNOT00000075725
|
RGD1566359
|
similar to RIKEN cDNA B230219D22 |
| chr6_-_135251073 | 3.84 |
ENSRNOT00000088986
|
Mok
|
MOK protein kinase |
| chr8_+_5676665 | 3.83 |
ENSRNOT00000012310
|
Mmp3
|
matrix metallopeptidase 3 |
| chr17_+_36334147 | 3.82 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chrX_-_158288109 | 3.68 |
ENSRNOT00000072527
|
LOC103690175
|
PHD finger protein 6 |
| chr3_-_66279155 | 3.67 |
ENSRNOT00000079887
|
Cerkl
|
ceramide kinase-like |
| chr2_-_149088787 | 3.66 |
ENSRNOT00000064833
|
Clrn1
|
clarin 1 |
| chr13_-_86671515 | 3.66 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr6_+_108285822 | 3.59 |
ENSRNOT00000015889
|
Vsx2
|
visual system homeobox 2 |
| chr1_+_282134981 | 3.45 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr5_-_152473868 | 3.43 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr5_+_107233230 | 3.34 |
ENSRNOT00000029976
|
Ifne
|
interferon, epsilon |
| chr1_+_263968134 | 3.33 |
ENSRNOT00000018090
|
LOC681458
|
similar to stearoyl-coenzyme A desaturase 3 |
| chr13_-_47440682 | 3.27 |
ENSRNOT00000037679
ENSRNOT00000005729 ENSRNOT00000050354 ENSRNOT00000050859 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr5_-_174921 | 3.20 |
ENSRNOT00000056239
|
Tipinl1
|
TIMELESS interacting protein like 1 |
| chr16_-_10802512 | 3.17 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr1_-_198900375 | 3.14 |
ENSRNOT00000024969
|
Zfp689
|
zinc finger protein 689 |
| chr7_+_132378273 | 3.14 |
ENSRNOT00000010990
|
LOC690142
|
hypothetical protein LOC690142 |
| chrX_+_11084317 | 3.11 |
ENSRNOT00000031808
ENSRNOT00000093745 |
RGD1565685
|
similar to RIKEN cDNA 1810030O07 |
| chr8_+_107508351 | 2.95 |
ENSRNOT00000081594
|
Cep70
|
centrosomal protein 70 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cpeb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.1 | 39.4 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 7.8 | 39.0 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 7.8 | 23.3 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 6.8 | 61.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 6.3 | 31.6 | GO:0061743 | motor learning(GO:0061743) |
| 5.8 | 29.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 5.7 | 17.0 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 3.7 | 22.3 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 3.6 | 14.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 3.4 | 10.3 | GO:0021627 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 3.4 | 13.4 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 3.3 | 13.0 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 3.2 | 3.2 | GO:0048378 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 3.1 | 9.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 3.1 | 12.3 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 3.0 | 11.9 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 3.0 | 80.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 2.8 | 8.4 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 2.7 | 8.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 2.7 | 38.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 2.6 | 7.8 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 2.6 | 7.7 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 2.5 | 7.4 | GO:1903165 | response to polycyclic arene(GO:1903165) |
| 2.4 | 7.2 | GO:1990743 | protein sialylation(GO:1990743) |
| 2.3 | 2.3 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 2.3 | 15.9 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 2.3 | 6.8 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 2.2 | 31.3 | GO:0007614 | short-term memory(GO:0007614) |
| 2.2 | 6.7 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 2.2 | 19.7 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 2.2 | 28.0 | GO:0071317 | cellular response to isoquinoline alkaloid(GO:0071317) |
| 2.1 | 37.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 2.0 | 35.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 1.9 | 5.7 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 1.9 | 11.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 1.8 | 7.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 1.8 | 12.3 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 1.7 | 6.8 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 1.7 | 34.8 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 1.6 | 24.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 1.6 | 23.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 1.6 | 11.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 1.6 | 19.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 1.6 | 16.2 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.6 | 6.3 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 1.5 | 6.2 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 1.5 | 8.8 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 1.5 | 16.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 1.4 | 15.0 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 1.4 | 9.5 | GO:0021902 | conditioned taste aversion(GO:0001661) commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.4 | 6.8 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 1.3 | 14.6 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 1.3 | 14.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 1.2 | 7.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 1.2 | 24.6 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 1.2 | 3.5 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 1.1 | 7.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.1 | 4.4 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 1.1 | 5.4 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 1.1 | 13.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 1.0 | 14.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 1.0 | 2.9 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.9 | 4.6 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.9 | 12.3 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.9 | 4.3 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.8 | 9.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.8 | 9.3 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.8 | 18.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.8 | 6.8 | GO:0035989 | tendon development(GO:0035989) |
| 0.8 | 21.2 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.7 | 8.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.7 | 2.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.7 | 28.5 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.7 | 4.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.7 | 25.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.7 | 2.7 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.7 | 6.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.6 | 8.1 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.6 | 16.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.6 | 2.3 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.6 | 1.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.5 | 3.8 | GO:0071460 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) cellular response to cell-matrix adhesion(GO:0071460) |
| 0.5 | 3.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.5 | 1.6 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.5 | 28.3 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.5 | 6.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.5 | 2.4 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.5 | 1.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.5 | 5.6 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.5 | 3.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.5 | 1.8 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.5 | 3.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.4 | 6.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.4 | 8.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.4 | 1.7 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.4 | 2.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.4 | 1.7 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 2.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.4 | 6.9 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.4 | 2.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.4 | 4.9 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.4 | 4.9 | GO:0060004 | reflex(GO:0060004) |
| 0.4 | 2.6 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.4 | 6.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.4 | 6.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 16.1 | GO:0090102 | cochlea development(GO:0090102) |
| 0.3 | 13.3 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.3 | 1.0 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.3 | 3.6 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 | 67.6 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.3 | 16.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.3 | 1.2 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.3 | 10.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.3 | 5.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.2 | 9.1 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.2 | 25.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 2.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 43.6 | GO:0016358 | dendrite development(GO:0016358) |
| 0.2 | 2.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.2 | 4.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 | 3.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 6.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 13.1 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 5.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 8.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 2.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 3.8 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.1 | 14.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 11.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 8.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 2.0 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 6.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.5 | GO:1902358 | sulfate transport(GO:0008272) oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.1 | 9.7 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.1 | 0.4 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 10.0 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.1 | 5.0 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.1 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 7.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 2.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 1.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 2.2 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 1.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 2.7 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 2.1 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.9 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 1.2 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.7 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 1.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.6 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 3.3 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 3.4 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 52.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 7.9 | 39.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 6.4 | 57.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 2.5 | 56.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 2.3 | 6.8 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 2.1 | 23.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 2.1 | 31.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.6 | 23.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.4 | 17.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.3 | 8.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.3 | 7.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.3 | 14.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 1.2 | 4.8 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 1.2 | 11.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 1.2 | 19.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 1.0 | 108.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 1.0 | 6.0 | GO:0033093 | Weibel-Palade body(GO:0033093) rough endoplasmic reticulum lumen(GO:0048237) |
| 0.9 | 11.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.9 | 22.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.8 | 5.8 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.8 | 16.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.8 | 10.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.7 | 18.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.6 | 6.0 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.6 | 8.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.6 | 11.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.5 | 7.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.5 | 6.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.5 | 1.6 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.5 | 2.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.5 | 13.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.5 | 2.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.4 | 6.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.4 | 6.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.4 | 16.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 14.6 | GO:0043034 | costamere(GO:0043034) |
| 0.4 | 40.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.4 | 4.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 15.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 1.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 3.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.3 | 4.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 1.8 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.3 | 4.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 56.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.3 | 6.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 5.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 35.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 24.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 1.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 10.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.2 | 5.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 3.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 45.9 | GO:0098793 | presynapse(GO:0098793) |
| 0.2 | 8.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 12.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 1.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 10.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 3.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 5.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 13.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 3.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 10.1 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 27.9 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 5.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 1.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 10.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 14.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 7.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 4.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.7 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 11.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 8.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 6.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.2 | GO:1902911 | protein kinase complex(GO:1902911) |
| 0.0 | 4.2 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 43.2 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.8 | GO:0005813 | centrosome(GO:0005813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 38.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 7.5 | 52.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 7.5 | 60.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 7.4 | 22.3 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 6.4 | 19.2 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 4.6 | 13.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 4.5 | 22.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 3.7 | 71.0 | GO:0030955 | potassium ion binding(GO:0030955) |
| 3.1 | 9.2 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 2.9 | 14.6 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 2.9 | 14.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 2.9 | 23.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 2.8 | 16.9 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 2.7 | 8.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 2.5 | 15.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 2.5 | 7.4 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 2.5 | 29.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 2.5 | 19.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 2.4 | 7.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 2.1 | 8.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 2.0 | 6.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 2.0 | 16.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 2.0 | 39.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 1.9 | 29.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 1.9 | 37.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 1.8 | 7.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.8 | 5.4 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 1.7 | 40.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 1.7 | 28.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.5 | 19.7 | GO:0019841 | retinol binding(GO:0019841) |
| 1.5 | 21.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 1.4 | 7.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.4 | 47.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.4 | 23.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 1.3 | 7.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 1.2 | 12.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.2 | 4.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 1.2 | 16.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 1.1 | 8.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.1 | 18.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 1.1 | 51.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 1.1 | 5.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 1.1 | 11.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.0 | 6.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 1.0 | 26.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.9 | 28.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.9 | 6.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.9 | 7.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.9 | 2.6 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.9 | 23.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.8 | 18.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.8 | 6.5 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.8 | 7.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.7 | 3.7 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.7 | 11.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.6 | 16.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.6 | 1.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.6 | 7.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.6 | 5.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.6 | 26.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.5 | 3.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.5 | 4.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.5 | 2.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 27.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.4 | 15.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.4 | 15.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.3 | 29.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.3 | 1.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.3 | 3.3 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.3 | 8.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 16.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 4.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 1.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 24.3 | GO:0070035 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.2 | 2.5 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.2 | 4.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 1.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 1.6 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.2 | 5.6 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.2 | 2.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 5.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 6.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.7 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 1.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 12.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 8.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 2.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 24.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 4.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 4.8 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 8.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 21.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 2.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.4 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 5.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 9.5 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 0.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.0 | 19.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 6.9 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 2.6 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 0.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.7 | GO:0051020 | GTPase binding(GO:0051020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 16.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 1.0 | 13.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.6 | 16.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.6 | 6.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.6 | 31.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.6 | 24.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.6 | 17.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.5 | 29.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 11.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 4.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 22.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.3 | 17.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.3 | 7.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.3 | 13.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.2 | 7.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 6.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 1.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 6.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 4.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 3.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 56.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 3.3 | 52.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 2.8 | 39.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.4 | 23.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 1.3 | 15.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 1.2 | 16.9 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 1.1 | 59.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 1.0 | 16.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.9 | 28.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.8 | 9.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.8 | 22.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.7 | 11.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.7 | 12.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.7 | 7.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.6 | 17.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.6 | 12.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.6 | 7.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.5 | 14.3 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.5 | 26.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 8.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.4 | 2.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.4 | 6.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 2.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 10.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 8.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 6.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 6.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.3 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 7.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 5.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 7.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 31.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.2 | 9.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 2.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 2.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 7.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 3.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 6.2 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.1 | 2.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.7 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 2.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |