Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Clock
Z-value: 0.42
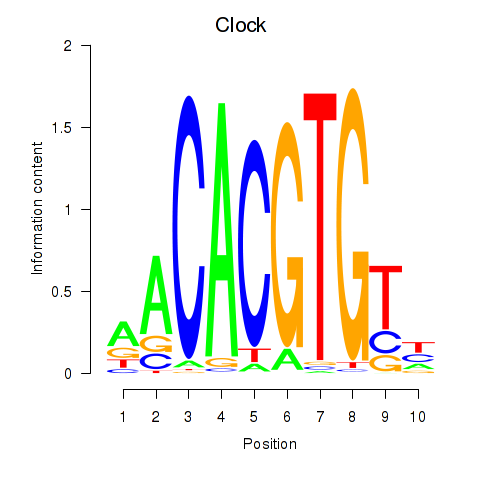
Transcription factors associated with Clock
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Clock
|
ENSRNOG00000002175 | clock circadian regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Clock | rn6_v1_chr14_+_34455934_34455934 | -0.09 | 1.1e-01 | Click! |
Activity profile of Clock motif
Sorted Z-values of Clock motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_83898527 | 12.72 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr9_-_82419288 | 11.28 |
ENSRNOT00000004797
|
Tuba4a
|
tubulin, alpha 4A |
| chr2_-_233743866 | 11.28 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr5_+_140979435 | 8.11 |
ENSRNOT00000056592
|
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr13_-_70783515 | 7.46 |
ENSRNOT00000003605
|
Lamc1
|
laminin subunit gamma 1 |
| chr2_-_30246010 | 7.27 |
ENSRNOT00000023900
|
Mccc2
|
methylcrotonoyl-CoA carboxylase 2 |
| chr15_+_4064706 | 7.10 |
ENSRNOT00000011956
|
Synpo2l
|
synaptopodin 2-like |
| chr10_-_32471454 | 6.78 |
ENSRNOT00000003224
|
Sgcd
|
sarcoglycan, delta |
| chr12_-_3924415 | 6.50 |
ENSRNOT00000067752
|
AABR07035074.1
|
|
| chr14_+_33580541 | 6.24 |
ENSRNOT00000002905
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr8_-_115140080 | 5.69 |
ENSRNOT00000015851
|
Acy1
|
aminoacylase 1 |
| chr6_-_55001464 | 5.56 |
ENSRNOT00000006618
|
Ahr
|
aryl hydrocarbon receptor |
| chr3_+_11424099 | 5.55 |
ENSRNOT00000019184
|
Ptges2
|
prostaglandin E synthase 2 |
| chrX_+_16946207 | 5.19 |
ENSRNOT00000003981
|
RGD1563606
|
similar to cysteine-rich protein 2 |
| chr3_+_113415774 | 5.08 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr3_-_163935617 | 4.90 |
ENSRNOT00000074023
|
Kcnb1
|
potassium voltage-gated channel subfamily B member 1 |
| chr5_+_167141875 | 4.80 |
ENSRNOT00000089314
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr10_+_83476107 | 4.69 |
ENSRNOT00000075733
|
Phb
|
prohibitin |
| chr1_+_219759183 | 4.59 |
ENSRNOT00000026316
|
Pc
|
pyruvate carboxylase |
| chr1_-_163328591 | 4.06 |
ENSRNOT00000034843
|
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr6_+_99282850 | 4.04 |
ENSRNOT00000008374
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1 |
| chr9_-_20621051 | 4.03 |
ENSRNOT00000015918
|
Tnfrsf21
|
TNF receptor superfamily member 21 |
| chr10_-_11760620 | 3.81 |
ENSRNOT00000009283
|
Dnase1
|
deoxyribonuclease 1 |
| chr10_-_62698541 | 3.79 |
ENSRNOT00000019759
|
Coro6
|
coronin 6 |
| chr6_+_80220547 | 3.51 |
ENSRNOT00000086221
ENSRNOT00000077435 ENSRNOT00000059318 ENSRNOT00000089010 |
Mia2
|
melanoma inhibitory activity 2 |
| chr7_+_123043503 | 3.51 |
ENSRNOT00000026258
ENSRNOT00000086355 |
Tef
|
TEF, PAR bZIP transcription factor |
| chr2_-_2683116 | 3.44 |
ENSRNOT00000016838
|
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr14_-_33580566 | 3.41 |
ENSRNOT00000063942
|
Paics
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr10_-_57618527 | 3.20 |
ENSRNOT00000037517
|
C1qbp
|
complement C1q binding protein |
| chr7_+_77966722 | 3.01 |
ENSRNOT00000006142
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr2_-_204032023 | 2.74 |
ENSRNOT00000040430
|
Atp1a1
|
ATPase Na+/K+ transporting subunit alpha 1 |
| chr10_-_37455022 | 2.70 |
ENSRNOT00000016742
|
LOC103694902
|
ubiquitin-conjugating enzyme E2 B |
| chr11_-_80826505 | 2.58 |
ENSRNOT00000032383
|
Rtp1
|
receptor (chemosensory) transporter protein 1 |
| chr16_-_75855745 | 2.54 |
ENSRNOT00000031291
|
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr4_+_78981987 | 2.50 |
ENSRNOT00000012490
|
Ccdc126
|
coiled-coil domain containing 126 |
| chr4_-_77489535 | 2.45 |
ENSRNOT00000008728
|
Pdia4
|
protein disulfide isomerase family A, member 4 |
| chr19_+_53055745 | 2.34 |
ENSRNOT00000074430
|
Foxl1
|
forkhead box L1 |
| chr6_+_43829945 | 2.19 |
ENSRNOT00000086548
|
Klf11
|
Kruppel-like factor 11 |
| chr11_-_83985385 | 1.93 |
ENSRNOT00000002334
|
LOC102551435
|
endothelin-converting enzyme 2-like |
| chrX_-_64702441 | 1.91 |
ENSRNOT00000051132
|
Amer1
|
APC membrane recruitment protein 1 |
| chr5_+_165405168 | 1.88 |
ENSRNOT00000014934
|
Srm
|
spermidine synthase |
| chr9_+_17817721 | 1.81 |
ENSRNOT00000086986
ENSRNOT00000026920 |
Hsp90ab1
|
heat shock protein 90 alpha family class B member 1 |
| chr7_-_27240528 | 1.74 |
ENSRNOT00000029435
ENSRNOT00000079731 |
Hsp90b1
|
heat shock protein 90 beta family member 1 |
| chr3_+_61685619 | 1.73 |
ENSRNOT00000002140
|
Hoxd1
|
homeo box D1 |
| chr11_+_47090245 | 1.72 |
ENSRNOT00000060645
|
AABR07033987.1
|
|
| chr8_+_44990014 | 1.63 |
ENSRNOT00000044608
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr10_+_46783979 | 1.45 |
ENSRNOT00000005237
|
Gid4
|
GID complex subunit 4 |
| chr4_-_78879294 | 1.44 |
ENSRNOT00000084543
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr5_-_58124681 | 1.34 |
ENSRNOT00000019795
|
Sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr10_-_40375605 | 1.33 |
ENSRNOT00000014464
|
Anxa6
|
annexin A6 |
| chr6_-_51257625 | 1.24 |
ENSRNOT00000012004
|
Hbp1
|
HMG-box transcription factor 1 |
| chr19_+_58565576 | 1.19 |
ENSRNOT00000026965
|
Ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr3_+_14588870 | 1.19 |
ENSRNOT00000071712
|
LOC100912604
|
spermidine synthase-like |
| chr14_+_36216002 | 1.18 |
ENSRNOT00000038506
|
Scfd2
|
sec1 family domain containing 2 |
| chr10_-_107324355 | 1.09 |
ENSRNOT00000064450
|
Usp36
|
ubiquitin specific peptidase 36 |
| chr3_+_148635775 | 1.03 |
ENSRNOT00000067261
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr2_-_148891900 | 0.99 |
ENSRNOT00000018488
|
Siah2
|
siah E3 ubiquitin protein ligase 2 |
| chr10_-_37209881 | 0.95 |
ENSRNOT00000090475
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr5_+_74649765 | 0.94 |
ENSRNOT00000075952
|
Palm2
|
paralemmin 2 |
| chrX_+_43881246 | 0.92 |
ENSRNOT00000005153
|
LOC317456
|
hypothetical LOC317456 |
| chr2_-_187160215 | 0.91 |
ENSRNOT00000092780
|
Ntrk1
|
neurotrophic receptor tyrosine kinase 1 |
| chr16_+_49462889 | 0.90 |
ENSRNOT00000039909
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr15_+_4554603 | 0.86 |
ENSRNOT00000075153
|
Kcnk5
|
potassium two pore domain channel subfamily K member 5 |
| chr10_+_88764732 | 0.84 |
ENSRNOT00000026662
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr7_-_83701447 | 0.82 |
ENSRNOT00000088893
|
AABR07057675.1
|
|
| chr13_-_90814119 | 0.80 |
ENSRNOT00000011208
|
Tagln2
|
transgelin 2 |
| chr4_+_113948514 | 0.75 |
ENSRNOT00000011521
|
LOC103692171
|
mannosyl-oligosaccharide glucosidase |
| chr1_-_198120061 | 0.70 |
ENSRNOT00000026231
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr6_+_33786627 | 0.69 |
ENSRNOT00000008359
|
Pum2
|
pumilio RNA-binding family member 2 |
| chr5_+_116421894 | 0.66 |
ENSRNOT00000080577
ENSRNOT00000086628 ENSRNOT00000004017 |
Nfia
|
nuclear factor I/A |
| chr13_+_35554964 | 0.51 |
ENSRNOT00000072632
|
Tmem185b
|
transmembrane protein 185B |
| chr1_+_198120099 | 0.45 |
ENSRNOT00000073652
|
Bola2
|
bolA family member 2 |
| chr16_+_7103998 | 0.29 |
ENSRNOT00000016581
|
Pbrm1
|
polybromo 1 |
| chr9_-_61690956 | 0.15 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr3_+_122803772 | 0.12 |
ENSRNOT00000009564
|
Nop56
|
NOP56 ribonucleoprotein |
| chr13_+_100129761 | 0.03 |
ENSRNOT00000038340
|
Eif3el1
|
eukaryotic translation initiation factor 3, subunit E-like 1 |
| chr10_+_59000397 | 0.02 |
ENSRNOT00000021134
|
Mybbp1a
|
MYB binding protein 1a |
| chr3_+_107142762 | 0.01 |
ENSRNOT00000006177
|
RGD1563680
|
similar to CDNA sequence BC052040 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Clock
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 2.3 | 11.3 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 2.1 | 6.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.4 | 5.6 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 1.3 | 4.0 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 1.2 | 4.7 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 1.1 | 4.6 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.1 | 3.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 1.0 | 4.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 1.0 | 4.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 1.0 | 4.8 | GO:0015755 | fructose transport(GO:0015755) |
| 0.8 | 4.9 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.8 | 4.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.8 | 2.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.8 | 3.0 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.7 | 3.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.6 | 3.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.6 | 1.8 | GO:0034486 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.6 | 3.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.5 | 2.7 | GO:0031947 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) potassium ion import across plasma membrane(GO:1990573) |
| 0.4 | 12.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.4 | 7.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.9 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.2 | 0.8 | GO:0060375 | development of secondary male sexual characteristics(GO:0046544) regulation of mast cell differentiation(GO:0060375) |
| 0.2 | 0.7 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.2 | 1.7 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 1.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 2.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 3.8 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 2.6 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.1 | 1.0 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 8.1 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.1 | 0.9 | GO:2000189 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 7.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.5 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.1 | 5.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.7 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 1.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 0.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.2 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.0 | 0.1 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 3.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 6.6 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 2.5 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.8 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 1.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 2.5 | GO:0006639 | acylglycerol metabolic process(GO:0006639) |
| 0.0 | 4.2 | GO:0006520 | cellular amino acid metabolic process(GO:0006520) |
| 0.0 | 1.0 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 1.5 | 7.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 1.1 | 6.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.0 | 12.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.9 | 4.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.8 | 11.3 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.5 | 1.6 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.4 | 4.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 0.7 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.2 | 2.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 3.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 8.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 4.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 7.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 6.1 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 3.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 12.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0090544 | BAF-type complex(GO:0090544) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 1.9 | 5.6 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 1.8 | 5.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 1.6 | 4.7 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 1.5 | 4.6 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.0 | 4.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.9 | 7.3 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.8 | 7.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.8 | 3.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.6 | 12.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.6 | 2.5 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.6 | 1.8 | GO:0017098 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.6 | 3.5 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.5 | 4.8 | GO:0070061 | fructose binding(GO:0070061) |
| 0.5 | 2.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.5 | 11.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.4 | 4.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.4 | 1.6 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.3 | 2.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 4.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 3.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 1.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 2.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 1.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.2 | 1.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 4.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 2.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.2 | 6.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.2 | 3.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 3.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 11.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.8 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 3.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 1.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 10.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 3.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 3.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 5.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 3.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.8 | 12.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.6 | 11.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 7.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 4.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 1.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 3.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 1.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 4.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 4.9 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 4.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 7.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 2.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.4 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |