Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
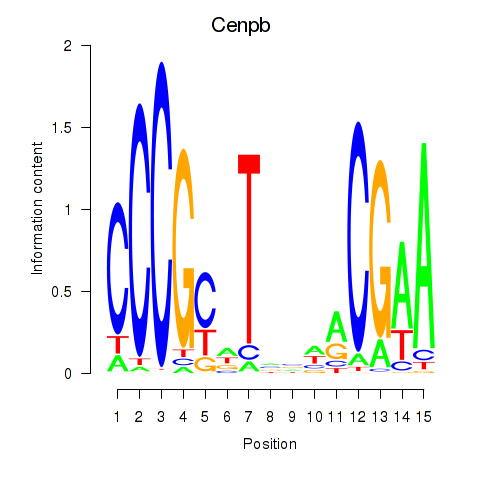
Results for Cenpb
Z-value: 0.65
Transcription factors associated with Cenpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cenpb
|
ENSRNOG00000057284 | centromere protein B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cenpb | rn6_v1_chr3_-_123723524_123723524 | 0.37 | 4.6e-12 | Click! |
Activity profile of Cenpb motif
Sorted Z-values of Cenpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_92530938 | 14.76 |
ENSRNOT00000064875
|
Slc16a14
|
solute carrier family 16, member 14 |
| chr16_-_32868680 | 13.36 |
ENSRNOT00000015974
ENSRNOT00000082392 |
Aadat
|
aminoadipate aminotransferase |
| chrX_+_74315850 | 12.95 |
ENSRNOT00000076538
|
LOC680227
|
LRRGT00193 |
| chr14_-_67170361 | 10.10 |
ENSRNOT00000005477
|
Slit2
|
slit guidance ligand 2 |
| chr1_-_165967069 | 9.76 |
ENSRNOT00000089359
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr11_+_86797557 | 9.42 |
ENSRNOT00000083049
ENSRNOT00000046594 |
Tango2
|
transport and golgi organization 2 homolog |
| chr9_+_82741920 | 8.86 |
ENSRNOT00000027337
|
Slc4a3
|
solute carrier family 4 member 3 |
| chr19_-_49532811 | 8.64 |
ENSRNOT00000015967
|
Gcsh
|
glycine cleavage system protein H |
| chr11_+_84396033 | 8.62 |
ENSRNOT00000002316
|
Abcc5
|
ATP binding cassette subfamily C member 5 |
| chrM_+_3904 | 8.56 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr20_-_4368693 | 8.30 |
ENSRNOT00000000501
|
Rnf5
|
ring finger protein 5 |
| chr7_+_13062196 | 8.30 |
ENSRNOT00000000193
|
Plpp2
|
phospholipid phosphatase 2 |
| chr9_-_82477136 | 8.26 |
ENSRNOT00000026753
ENSRNOT00000089555 |
Resp18
|
regulated endocrine-specific protein 18 |
| chr3_-_151106557 | 8.02 |
ENSRNOT00000025657
|
Gss
|
glutathione synthetase |
| chr15_-_41338284 | 7.93 |
ENSRNOT00000077225
|
Tnfrsf19
|
TNF receptor superfamily member 19 |
| chr10_+_56824505 | 7.55 |
ENSRNOT00000067128
|
Slc16a11
|
solute carrier family 16, member 11 |
| chr14_-_85191557 | 7.44 |
ENSRNOT00000011604
|
Nefh
|
neurofilament heavy |
| chr6_+_43234526 | 6.86 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr7_-_116284587 | 6.84 |
ENSRNOT00000073179
|
LOC100910656
|
ly-6/neurotoxin-like protein 1-like |
| chr7_+_3320103 | 6.81 |
ENSRNOT00000090381
|
Cd63
|
Cd63 molecule |
| chr20_-_48758905 | 6.79 |
ENSRNOT00000000710
|
Ddo
|
D-aspartate oxidase |
| chr16_-_21348391 | 6.78 |
ENSRNOT00000083537
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr12_-_22417980 | 6.76 |
ENSRNOT00000072208
|
Ephb4
|
EPH receptor B4 |
| chr4_+_89079014 | 6.30 |
ENSRNOT00000087451
|
Herc3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr2_-_26011429 | 6.20 |
ENSRNOT00000065143
|
Arhgef28
|
Rho guanine nucleotide exchange factor 28 |
| chr9_-_4168221 | 6.16 |
ENSRNOT00000061861
|
Sult1c2a
|
sulfotransferase family, cytosolic, 1C, member 2a |
| chr8_-_55087832 | 6.15 |
ENSRNOT00000032152
|
Dlat
|
dihydrolipoamide S-acetyltransferase |
| chr12_+_12374790 | 6.06 |
ENSRNOT00000001347
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chr9_+_94362299 | 5.98 |
ENSRNOT00000021168
|
Efhd1
|
EF-hand domain family, member D1 |
| chr2_-_9504134 | 5.90 |
ENSRNOT00000076996
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chrX_+_75382598 | 5.81 |
ENSRNOT00000033494
|
Uprt
|
uracil phosphoribosyltransferase homolog |
| chr19_-_42920344 | 5.80 |
ENSRNOT00000019408
ENSRNOT00000068594 |
Zfhx3
|
zinc finger homeobox 3 |
| chrX_-_105622156 | 5.58 |
ENSRNOT00000029511
|
Armcx2
|
armadillo repeat containing, X-linked 2 |
| chr3_-_52664209 | 5.57 |
ENSRNOT00000065126
ENSRNOT00000079020 |
Scn9a
|
sodium voltage-gated channel alpha subunit 9 |
| chr1_-_216663720 | 5.53 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr19_-_58399816 | 5.38 |
ENSRNOT00000026843
|
Sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr14_+_34727623 | 5.36 |
ENSRNOT00000071405
ENSRNOT00000090183 |
Kdr
|
kinase insert domain receptor |
| chr2_+_178679041 | 5.23 |
ENSRNOT00000013524
|
Fam198b
|
family with sequence similarity 198, member B |
| chr16_-_73410777 | 5.22 |
ENSRNOT00000024128
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr3_+_139894331 | 5.21 |
ENSRNOT00000064695
|
Rin2
|
Ras and Rab interactor 2 |
| chr1_+_67025240 | 5.21 |
ENSRNOT00000051024
|
Vom1r47
|
vomeronasal 1 receptor 47 |
| chr3_-_146470293 | 5.16 |
ENSRNOT00000009627
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr15_+_42960307 | 5.09 |
ENSRNOT00000012528
|
Trim35
|
tripartite motif-containing 35 |
| chr9_+_47281961 | 5.08 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr14_-_10395047 | 5.00 |
ENSRNOT00000002936
|
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr19_+_61332351 | 4.87 |
ENSRNOT00000014492
|
Nrp1
|
neuropilin 1 |
| chr10_+_94260197 | 4.86 |
ENSRNOT00000063973
|
Taco1
|
translational activator of cytochrome c oxidase I |
| chr8_-_122987191 | 4.82 |
ENSRNOT00000033976
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr14_+_34727915 | 4.76 |
ENSRNOT00000085089
|
Kdr
|
kinase insert domain receptor |
| chr20_-_14573519 | 4.75 |
ENSRNOT00000001772
|
Rab36
|
RAB36, member RAS oncogene family |
| chr12_-_24046814 | 4.72 |
ENSRNOT00000001961
|
Por
|
cytochrome p450 oxidoreductase |
| chr1_+_220416018 | 4.56 |
ENSRNOT00000027233
|
B4gat1
|
beta-1,4-glucuronyltransferase 1 |
| chr1_-_128695995 | 4.54 |
ENSRNOT00000077020
|
Synm
|
synemin |
| chr1_+_80383050 | 4.54 |
ENSRNOT00000023451
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr7_+_125034764 | 4.48 |
ENSRNOT00000015767
|
Pnpla3
|
patatin-like phospholipase domain containing 3 |
| chr3_+_7053528 | 4.45 |
ENSRNOT00000082883
|
Mrps2
|
mitochondrial ribosomal protein S2 |
| chr3_-_160891190 | 4.43 |
ENSRNOT00000019386
|
Sdc4
|
syndecan 4 |
| chr2_-_235177275 | 4.39 |
ENSRNOT00000093153
|
LOC103691699
|
uncharacterized LOC103691699 |
| chrX_-_56765893 | 4.35 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr13_-_107831014 | 4.33 |
ENSRNOT00000003684
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr13_-_80819218 | 4.30 |
ENSRNOT00000072922
|
Fmo6
|
flavin containing monooxygenase 6 |
| chr9_+_66568074 | 4.27 |
ENSRNOT00000035238
|
Bmpr2
|
bone morphogenetic protein receptor type 2 |
| chr4_-_100303047 | 4.23 |
ENSRNOT00000018170
ENSRNOT00000084782 |
Mat2a
|
methionine adenosyltransferase 2A |
| chr8_+_49354115 | 4.18 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr20_+_3552929 | 4.14 |
ENSRNOT00000083223
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr5_+_147069616 | 4.14 |
ENSRNOT00000072908
|
Trim62
|
tripartite motif-containing 62 |
| chr14_-_112946875 | 4.11 |
ENSRNOT00000081981
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr17_+_78735598 | 4.02 |
ENSRNOT00000020854
|
Hspa14
|
heat shock protein family A, member 14 |
| chr7_-_130101858 | 4.01 |
ENSRNOT00000007954
|
Tubgcp6
|
tubulin, gamma complex associated protein 6 |
| chr4_+_104949597 | 4.01 |
ENSRNOT00000073104
|
LOC100910130
|
protein FAM50A-like |
| chr15_-_28721127 | 3.98 |
ENSRNOT00000017720
|
Mettl3
|
methyltransferase-like 3 |
| chr20_+_31313018 | 3.98 |
ENSRNOT00000036719
|
Tysnd1
|
trypsin domain containing 1 |
| chr6_+_136380990 | 3.95 |
ENSRNOT00000063865
ENSRNOT00000082914 |
Zfyve21
|
zinc finger FYVE-type containing 21 |
| chr12_+_29921443 | 3.93 |
ENSRNOT00000001190
|
Sbds
|
SBDS ribosome assembly guanine nucleotide exchange factor |
| chr1_+_220322940 | 3.92 |
ENSRNOT00000074972
|
LOC108348085
|
beta-1,4-glucuronyltransferase 1 |
| chr10_-_103816287 | 3.85 |
ENSRNOT00000004477
|
Grin2c
|
glutamate ionotropic receptor NMDA type subunit 2C |
| chr20_+_55594676 | 3.81 |
ENSRNOT00000057010
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chr1_-_18058055 | 3.80 |
ENSRNOT00000020988
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr15_+_87886783 | 3.78 |
ENSRNOT00000065710
|
Slain1
|
SLAIN motif family, member 1 |
| chr8_+_48699769 | 3.78 |
ENSRNOT00000044613
|
Hyou1
|
hypoxia up-regulated 1 |
| chr3_-_168111920 | 3.75 |
ENSRNOT00000046011
|
Cyp24a1
|
cytochrome P450, family 24, subfamily a, polypeptide 1 |
| chr9_-_50884596 | 3.73 |
ENSRNOT00000016285
|
Kdelc1
|
KDEL motif containing 1 |
| chr3_-_82856171 | 3.66 |
ENSRNOT00000088555
|
RGD1564664
|
similar to LOC387763 protein |
| chr11_+_87677966 | 3.59 |
ENSRNOT00000068711
|
Klhl22
|
kelch-like family member 22 |
| chr5_-_1347921 | 3.58 |
ENSRNOT00000007994
|
Gdap1
|
ganglioside-induced differentiation-associated-protein 1 |
| chr14_-_86868598 | 3.52 |
ENSRNOT00000087212
|
Nacad
|
NAC alpha domain containing |
| chr7_-_122926336 | 3.50 |
ENSRNOT00000000205
|
Chadl
|
chondroadherin-like |
| chr9_-_104350308 | 3.50 |
ENSRNOT00000033958
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr20_-_13667333 | 3.42 |
ENSRNOT00000031472
|
LOC103694872
|
coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial |
| chr18_+_1971506 | 3.41 |
ENSRNOT00000092027
|
Mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr4_+_81311490 | 3.40 |
ENSRNOT00000016265
|
Snx10
|
sorting nexin 10 |
| chr2_-_53827175 | 3.34 |
ENSRNOT00000078158
|
RGD1305938
|
similar to expressed sequence AW549877 |
| chr15_-_38276159 | 3.33 |
ENSRNOT00000084698
ENSRNOT00000015233 |
Micu2
|
mitochondrial calcium uptake 2 |
| chr1_-_87221826 | 3.29 |
ENSRNOT00000046611
ENSRNOT00000028006 |
Spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr16_-_21089508 | 3.26 |
ENSRNOT00000072565
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr19_+_56024903 | 3.22 |
ENSRNOT00000021607
|
Cdk10
|
cyclin-dependent kinase 10 |
| chr10_-_47453442 | 3.22 |
ENSRNOT00000050061
|
Usp22
|
ubiquitin specific peptidase 22 |
| chr10_-_57618527 | 3.17 |
ENSRNOT00000037517
|
C1qbp
|
complement C1q binding protein |
| chr8_-_69164758 | 3.13 |
ENSRNOT00000013933
|
Map2k1
|
mitogen activated protein kinase kinase 1 |
| chr3_-_3824284 | 3.12 |
ENSRNOT00000068110
ENSRNOT00000061682 |
Snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chrX_+_73390903 | 3.12 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chr1_+_88457594 | 3.08 |
ENSRNOT00000049877
|
Zfp260
|
zinc finger protein 260 |
| chr17_+_19249952 | 2.98 |
ENSRNOT00000023140
|
Atxn1
|
ataxin 1 |
| chr14_+_77380262 | 2.97 |
ENSRNOT00000008030
|
Nsg1
|
neuron specific gene family member 1 |
| chr1_-_101809544 | 2.90 |
ENSRNOT00000028591
|
Kcnj14
|
potassium voltage-gated channel subfamily J member 14 |
| chr9_-_94601852 | 2.85 |
ENSRNOT00000022485
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chrX_-_77061604 | 2.82 |
ENSRNOT00000083643
ENSRNOT00000089386 |
Magt1
|
magnesium transporter 1 |
| chr1_-_198120061 | 2.81 |
ENSRNOT00000026231
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr7_-_2353875 | 2.80 |
ENSRNOT00000074873
|
LOC100911319
|
zinc finger protein 36, C3H1 type-like 2-like |
| chr2_-_219628997 | 2.80 |
ENSRNOT00000064484
|
Trmt13
|
tRNA methyltransferase 13 homolog |
| chr1_+_87790104 | 2.77 |
ENSRNOT00000074580
|
LOC103690114
|
zinc finger protein 383-like |
| chrX_+_39711201 | 2.76 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr5_+_70245843 | 2.76 |
ENSRNOT00000083983
ENSRNOT00000082108 |
Slc44a1
|
solute carrier family 44 member 1 |
| chr11_+_87678295 | 2.72 |
ENSRNOT00000091675
|
Klhl22
|
kelch-like family member 22 |
| chr5_-_160070916 | 2.70 |
ENSRNOT00000067517
ENSRNOT00000055791 |
Spen
|
spen family transcriptional repressor |
| chr5_+_126783061 | 2.70 |
ENSRNOT00000013224
|
Tmem59
|
transmembrane protein 59 |
| chr5_+_63192298 | 2.69 |
ENSRNOT00000008329
|
Sec61b
|
Sec61 translocon beta subunit |
| chr1_+_115975324 | 2.68 |
ENSRNOT00000080907
|
Atp10a
|
ATPase phospholipid transporting 10A (putative) |
| chr8_-_113675128 | 2.66 |
ENSRNOT00000017318
|
Nudt16
|
nudix hydrolase 16 |
| chr3_+_8701855 | 2.65 |
ENSRNOT00000021431
|
Tbc1d13
|
TBC1 domain family, member 13 |
| chr1_-_219519398 | 2.65 |
ENSRNOT00000025546
|
Ssh3
|
slingshot protein phosphatase 3 |
| chr10_+_89578212 | 2.65 |
ENSRNOT00000028178
|
Arl4d
|
ADP-ribosylation factor like GTPase 4D |
| chr8_-_108880879 | 2.62 |
ENSRNOT00000020610
|
Slc35g2
|
solute carrier family 35, member G2 |
| chr20_+_4369178 | 2.58 |
ENSRNOT00000088079
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr10_-_108473377 | 2.57 |
ENSRNOT00000070934
|
Sgsh
|
N-sulfoglucosamine sulfohydrolase |
| chrX_-_1784807 | 2.57 |
ENSRNOT00000077453
|
Rbm10
|
RNA binding motif protein 10 |
| chr5_-_119564846 | 2.56 |
ENSRNOT00000012977
|
Cyp2j4
|
cytochrome P450, family 2, subfamily j, polypeptide 4 |
| chr1_-_225014538 | 2.56 |
ENSRNOT00000026376
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr3_-_113091770 | 2.56 |
ENSRNOT00000068132
|
Lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr3_-_3798177 | 2.55 |
ENSRNOT00000025386
ENSRNOT00000076156 |
Dnlz
|
DNL-type zinc finger |
| chr12_-_39699088 | 2.52 |
ENSRNOT00000088905
ENSRNOT00000001717 |
Vps29
|
VPS29 retromer complex component |
| chr10_-_94681914 | 2.51 |
ENSRNOT00000085736
|
Ern1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr8_-_59077690 | 2.49 |
ENSRNOT00000066197
|
Dmxl2
|
Dmx-like 2 |
| chr13_-_102942863 | 2.44 |
ENSRNOT00000003198
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr3_-_170955399 | 2.43 |
ENSRNOT00000084990
|
Bmp7
|
bone morphogenetic protein 7 |
| chr14_-_84978255 | 2.43 |
ENSRNOT00000078179
|
Cabp7
|
calcium binding protein 7 |
| chr6_+_48866601 | 2.42 |
ENSRNOT00000077321
ENSRNOT00000079891 |
Pxdn
|
peroxidasin |
| chr1_+_59765835 | 2.40 |
ENSRNOT00000014842
|
Fpr1
|
formyl peptide receptor 1 |
| chr12_-_41485122 | 2.40 |
ENSRNOT00000001859
|
Ddx54
|
DEAD-box helicase 54 |
| chr7_+_12532895 | 2.39 |
ENSRNOT00000018169
ENSRNOT00000093599 ENSRNOT00000093413 |
Polr2e
|
RNA polymerase II subunit E |
| chr3_-_5975734 | 2.38 |
ENSRNOT00000081376
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr18_-_2183508 | 2.35 |
ENSRNOT00000073306
|
AABR07031158.1
|
|
| chr10_+_4719713 | 2.35 |
ENSRNOT00000003412
|
Litaf
|
lipopolysaccharide-induced TNF factor |
| chr15_+_34352914 | 2.33 |
ENSRNOT00000067150
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr16_-_2367253 | 2.31 |
ENSRNOT00000083573
|
Pde12
|
phosphodiesterase 12 |
| chr10_-_91500661 | 2.31 |
ENSRNOT00000037147
|
Plekhm1
|
pleckstrin homology and RUN domain containing M1 |
| chr4_+_153876149 | 2.29 |
ENSRNOT00000018083
|
Slc6a13
|
solute carrier family 6 member 13 |
| chr3_-_77483322 | 2.29 |
ENSRNOT00000008077
|
Olr661
|
olfactory receptor 661 |
| chr8_-_50277797 | 2.29 |
ENSRNOT00000082508
|
Pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr18_+_29960072 | 2.28 |
ENSRNOT00000071366
|
AC103179.1
|
|
| chr4_-_150506406 | 2.27 |
ENSRNOT00000076307
|
Zfp248
|
zinc finger protein 248 |
| chr10_-_91186054 | 2.26 |
ENSRNOT00000004140
|
Plcd3
|
phospholipase C, delta 3 |
| chr1_+_36320461 | 2.25 |
ENSRNOT00000023659
|
Srd5a1
|
steroid 5 alpha-reductase 1 |
| chr20_-_29184185 | 2.24 |
ENSRNOT00000090771
|
Mcu
|
mitochondrial calcium uniporter |
| chr13_+_99221013 | 2.23 |
ENSRNOT00000083608
|
Tmem63a
|
transmembrane protein 63a |
| chr4_+_167219728 | 2.23 |
ENSRNOT00000075273
|
Smim10l1
|
small integral membrane protein 10 like 1 |
| chr6_-_98606109 | 2.23 |
ENSRNOT00000006860
|
Wdr89
|
WD repeat domain 89 |
| chr1_+_240908483 | 2.21 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
| chr10_-_90151042 | 2.21 |
ENSRNOT00000055187
|
Hdac5
|
histone deacetylase 5 |
| chr1_+_277355619 | 2.17 |
ENSRNOT00000022788
|
Nhlrc2
|
NHL repeat containing 2 |
| chr1_-_218810118 | 2.14 |
ENSRNOT00000065950
ENSRNOT00000020886 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chrX_+_76786466 | 2.14 |
ENSRNOT00000090665
|
Fgf16
|
fibroblast growth factor 16 |
| chr8_+_103774358 | 2.12 |
ENSRNOT00000014481
|
Xrn1
|
5'-3' exoribonuclease 1 |
| chr4_-_150506557 | 2.09 |
ENSRNOT00000076927
|
Zfp248
|
zinc finger protein 248 |
| chr18_-_73873280 | 2.09 |
ENSRNOT00000075580
|
Rnf165
|
ring finger protein 165 |
| chr1_-_46329682 | 2.09 |
ENSRNOT00000046311
|
Tmem242
|
transmembrane protein 242 |
| chr16_-_81583374 | 2.09 |
ENSRNOT00000092501
|
LOC103693999
|
transmembrane and coiled-coil domain-containing protein 3 |
| chr1_+_265761738 | 2.00 |
ENSRNOT00000024898
|
Hps6
|
Hermansky-Pudlak syndrome 6 |
| chr9_-_43127887 | 1.98 |
ENSRNOT00000021685
|
Ankrd39
|
ankyrin repeat domain 39 |
| chr16_-_2367069 | 1.98 |
ENSRNOT00000079625
|
Pde12
|
phosphodiesterase 12 |
| chr17_+_42241159 | 1.96 |
ENSRNOT00000024878
|
Acot13
|
acyl-CoA thioesterase 13 |
| chr8_+_116324040 | 1.95 |
ENSRNOT00000081353
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr11_+_73693814 | 1.95 |
ENSRNOT00000081081
ENSRNOT00000002354 ENSRNOT00000090940 |
Lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr4_+_148139528 | 1.92 |
ENSRNOT00000092594
ENSRNOT00000015620 ENSRNOT00000092613 |
Washc2c
|
WASH complex subunit 2C |
| chr19_+_41932221 | 1.87 |
ENSRNOT00000059225
|
LOC103694328
|
alanine and glycine-rich protein-like |
| chr10_+_89181180 | 1.87 |
ENSRNOT00000078931
ENSRNOT00000027770 |
Wnk4
|
WNK lysine deficient protein kinase 4 |
| chr1_-_198831691 | 1.86 |
ENSRNOT00000074435
|
Zfp764
|
zinc finger protein 764 |
| chr13_+_85465792 | 1.86 |
ENSRNOT00000082991
ENSRNOT00000005277 |
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr7_+_26256459 | 1.84 |
ENSRNOT00000010986
|
Appl2
|
adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 2 |
| chr15_-_4035064 | 1.82 |
ENSRNOT00000012300
|
Fut11
|
fucosyltransferase 11 |
| chr14_+_10581136 | 1.81 |
ENSRNOT00000002987
|
Coq2
|
coenzyme Q2, polyprenyltransferase |
| chr3_+_119698652 | 1.79 |
ENSRNOT00000088834
ENSRNOT00000066114 |
Stard7
|
StAR-related lipid transfer domain containing 7 |
| chr1_-_220096319 | 1.76 |
ENSRNOT00000091787
ENSRNOT00000073983 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr7_-_98098268 | 1.76 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr11_+_60907015 | 1.75 |
ENSRNOT00000002797
|
Gtpbp8
|
GTP-binding protein 8 (putative) |
| chr14_+_82769642 | 1.74 |
ENSRNOT00000065393
|
Ctbp1
|
C-terminal binding protein 1 |
| chr12_+_30514112 | 1.74 |
ENSRNOT00000001228
|
Psph
|
phosphoserine phosphatase |
| chr1_+_7101715 | 1.73 |
ENSRNOT00000019994
|
Sf3b5
|
splicing factor 3b, subunit 5 |
| chr1_-_204855627 | 1.73 |
ENSRNOT00000023121
|
Mettl10
|
methyltransferase like 10 |
| chr4_-_82702429 | 1.73 |
ENSRNOT00000011069
|
Hibadh
|
3-hydroxyisobutyrate dehydrogenase |
| chr8_+_62341613 | 1.72 |
ENSRNOT00000066923
|
Scamp2
|
secretory carrier membrane protein 2 |
| chr6_+_86785771 | 1.71 |
ENSRNOT00000066702
|
Prpf39
|
pre-mRNA processing factor 39 |
| chr1_-_214844858 | 1.69 |
ENSRNOT00000046344
|
Tollip
|
toll interacting protein |
| chr7_-_142132173 | 1.69 |
ENSRNOT00000026613
|
Csrnp2
|
cysteine and serine rich nuclear protein 2 |
| chr9_-_53097979 | 1.68 |
ENSRNOT00000005408
|
Osgepl1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr4_+_147756553 | 1.66 |
ENSRNOT00000086549
ENSRNOT00000014733 |
Ift122
|
intraflagellar transport 122 |
| chr1_-_225035687 | 1.65 |
ENSRNOT00000026539
|
Gng3
|
G protein subunit gamma 3 |
| chr1_-_236501733 | 1.65 |
ENSRNOT00000030491
|
Rfk
|
riboflavin kinase |
| chr15_+_33677597 | 1.63 |
ENSRNOT00000024337
|
Ngdn
|
neuroguidin |
| chr3_+_176821652 | 1.62 |
ENSRNOT00000055030
|
Rtel1
|
regulator of telomere elongation helicase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cenpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.4 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 3.4 | 10.1 | GO:0071623 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 2.5 | 7.4 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 2.4 | 2.4 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 2.3 | 6.8 | GO:0006533 | aspartate catabolic process(GO:0006533) D-amino acid metabolic process(GO:0046416) |
| 2.2 | 8.6 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.8 | 7.3 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 1.7 | 5.2 | GO:0071504 | cellular response to heparin(GO:0071504) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 1.7 | 5.2 | GO:0019541 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 1.7 | 10.1 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 1.6 | 4.9 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) renal artery morphogenesis(GO:0061441) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 1.6 | 4.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.4 | 5.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 1.4 | 4.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 1.4 | 4.3 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) regulation of lung blood pressure(GO:0014916) negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 1.4 | 5.6 | GO:0043179 | rhythmic excitation(GO:0043179) |
| 1.3 | 6.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 1.3 | 3.9 | GO:0033058 | directional locomotion(GO:0033058) |
| 1.2 | 3.7 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 1.2 | 4.7 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) nitrate metabolic process(GO:0042126) |
| 1.1 | 4.4 | GO:1903553 | aortic smooth muscle cell differentiation(GO:0035887) positive regulation of extracellular exosome assembly(GO:1903553) |
| 1.1 | 3.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 1.0 | 5.8 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.9 | 5.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.9 | 4.6 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.9 | 2.7 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.9 | 2.7 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.9 | 6.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.9 | 2.6 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.9 | 6.8 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.8 | 4.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.8 | 5.9 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.8 | 3.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.8 | 5.8 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.8 | 1.6 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.8 | 3.3 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.8 | 0.8 | GO:0021888 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.8 | 3.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.7 | 2.2 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.7 | 2.8 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.7 | 8.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.7 | 3.9 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.6 | 1.9 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.6 | 8.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.6 | 1.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.6 | 1.7 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.6 | 1.7 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.6 | 5.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.5 | 1.6 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.5 | 1.6 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.5 | 3.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.5 | 4.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.5 | 2.5 | GO:0030969 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.5 | 2.0 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.5 | 3.0 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.5 | 1.9 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.5 | 3.8 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.5 | 1.4 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.5 | 1.4 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.5 | 9.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.5 | 1.4 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.4 | 1.3 | GO:0042245 | RNA repair(GO:0042245) |
| 0.4 | 6.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.4 | 1.7 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.4 | 4.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.4 | 1.3 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.4 | 2.1 | GO:1904849 | positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.4 | 1.7 | GO:0060830 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.4 | 1.6 | GO:1904429 | regulation of t-circle formation(GO:1904429) negative regulation of t-circle formation(GO:1904430) |
| 0.4 | 2.0 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.4 | 5.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 4.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.4 | 1.5 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.4 | 0.7 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.4 | 1.8 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.4 | 3.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.3 | 1.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 2.8 | GO:0015871 | choline transport(GO:0015871) |
| 0.3 | 1.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.3 | 0.7 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.3 | 1.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.3 | 2.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.3 | 1.2 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.3 | 1.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 0.6 | GO:1903751 | negative regulation of cellular respiration(GO:1901856) regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.3 | 1.1 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.3 | 0.8 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.3 | 1.4 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 | 3.8 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.3 | 3.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 8.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 0.8 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.3 | 1.0 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.3 | 5.1 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.3 | 1.5 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 2.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 3.3 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 | 0.9 | GO:0021586 | pons maturation(GO:0021586) |
| 0.2 | 1.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 8.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 1.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 6.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 5.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 2.9 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.2 | 0.6 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 2.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.4 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 4.0 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.2 | 1.6 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 1.2 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.2 | 2.5 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 6.0 | GO:0071174 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.2 | 0.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 1.9 | GO:0006983 | ER overload response(GO:0006983) |
| 0.2 | 2.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 4.1 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.2 | 1.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 2.3 | GO:0042448 | progesterone metabolic process(GO:0042448) circadian sleep/wake cycle, REM sleep(GO:0042747) |
| 0.2 | 1.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 5.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 2.6 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.2 | 1.5 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.2 | 2.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 0.5 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.2 | 4.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.2 | 1.4 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.2 | 3.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 2.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 1.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.6 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 3.6 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.4 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.6 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 1.8 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 1.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.0 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 3.9 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 4.3 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.8 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 1.8 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 3.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 1.7 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 2.3 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.1 | 2.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 2.3 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 2.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 1.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 1.4 | GO:0031498 | chromatin disassembly(GO:0031498) |
| 0.1 | 2.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 0.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.6 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 1.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 1.8 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 0.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.7 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 9.7 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.1 | 1.4 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 2.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 2.6 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 1.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.1 | 1.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.4 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 1.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 2.1 | GO:0060384 | innervation(GO:0060384) |
| 0.1 | 2.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 2.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.6 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 1.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 4.6 | GO:0001657 | ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.1 | 1.0 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 1.6 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 4.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.5 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.4 | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 1.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 2.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 2.2 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 2.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.3 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 1.1 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 4.6 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.4 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.2 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 2.0 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 2.4 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.8 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 8.2 | GO:0030036 | actin cytoskeleton organization(GO:0030036) |
| 0.0 | 1.3 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.6 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.0 | GO:0016128 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.0 | 1.0 | GO:0006664 | glycolipid metabolic process(GO:0006664) |
| 0.0 | 2.1 | GO:0032868 | response to insulin(GO:0032868) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 1.9 | 15.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.7 | 6.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.4 | 4.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.4 | 8.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.2 | 5.8 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 1.0 | 5.9 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 1.0 | 4.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.9 | 2.8 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.9 | 5.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.9 | 6.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.9 | 2.7 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.8 | 4.2 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.8 | 2.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.7 | 7.4 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.7 | 4.0 | GO:0036396 | MIS complex(GO:0036396) |
| 0.6 | 8.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.6 | 2.5 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.6 | 2.5 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.6 | 6.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.5 | 4.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.5 | 1.5 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.4 | 2.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.4 | 1.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.4 | 3.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 1.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 3.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 1.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.3 | 2.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 0.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 5.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 1.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 1.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.3 | 0.8 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 1.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 1.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 3.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 1.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 5.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 2.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 2.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 8.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 2.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 1.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 4.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 4.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.0 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.1 | 2.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.6 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 1.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 2.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 4.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 4.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.6 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 3.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 2.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.8 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 5.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 5.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 1.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.3 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 6.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 10.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 1.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 7.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.8 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.0 | 3.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 42.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 29.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 13.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 4.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.9 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.4 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 1.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.4 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 2.5 | 10.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 1.9 | 15.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 1.4 | 4.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 1.3 | 4.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 1.3 | 5.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.2 | 5.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 1.1 | 4.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.0 | 4.8 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.9 | 4.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.9 | 2.6 | GO:0008405 | arachidonic acid 11,12-epoxygenase activity(GO:0008405) |
| 0.8 | 5.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.8 | 8.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.8 | 3.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.8 | 2.3 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.8 | 6.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.8 | 8.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.7 | 2.2 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.7 | 4.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.7 | 2.7 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.7 | 2.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.6 | 4.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.6 | 4.7 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.6 | 1.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.6 | 2.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.6 | 2.3 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.6 | 4.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.6 | 2.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.6 | 2.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.5 | 2.7 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.5 | 1.6 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.5 | 3.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.5 | 5.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 1.5 | GO:0005128 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 0.5 | 3.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 1.4 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.5 | 1.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.4 | 8.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.4 | 7.3 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.4 | 1.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.4 | 2.8 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.4 | 3.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 2.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.4 | 3.0 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.4 | 7.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.4 | 5.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.4 | 4.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.4 | 4.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 1.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 2.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.4 | 1.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 9.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 1.3 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.3 | 3.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 1.8 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 2.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.3 | 6.8 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) FAD binding(GO:0071949) |
| 0.3 | 1.7 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 4.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 3.7 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.3 | 1.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.3 | 3.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 3.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 8.9 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.2 | 5.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.2 | 0.7 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 8.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 2.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.2 | 1.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 2.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 5.6 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.2 | 2.7 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 2.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 2.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 5.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 3.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 1.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 1.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 4.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 1.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 1.7 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.2 | 0.8 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 3.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 2.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 2.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.5 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 1.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 4.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 2.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.8 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 1.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 2.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 6.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 1.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 2.0 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 5.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 3.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 3.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 2.4 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 2.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 1.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 5.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 11.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 6.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.3 | GO:0016675 | oxidoreductase activity, acting on a heme group of donors(GO:0016675) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 5.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 4.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 1.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 4.9 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 1.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 2.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 11.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 1.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.4 | GO:0015605 | nucleotide transmembrane transporter activity(GO:0015215) organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.0 | 1.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.6 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.6 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 15.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.5 | 6.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 10.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 6.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 7.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 0.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.2 | 1.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 10.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 3.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 6.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 8.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 4.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 7.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 5.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 6.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 3.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 3.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 7.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 2.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 13.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 1.2 | 15.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.6 | 5.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 10.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.5 | 10.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 7.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.4 | 6.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 3.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.3 | 4.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 4.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 7.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 5.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 2.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 3.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 1.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 2.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 8.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 4.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 2.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 4.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 3.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 5.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 2.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 4.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 6.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 4.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 1.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 4.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 7.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 9.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 2.2 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.4 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.2 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |