Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Cebpa_Cebpg
Z-value: 1.54
Transcription factors associated with Cebpa_Cebpg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
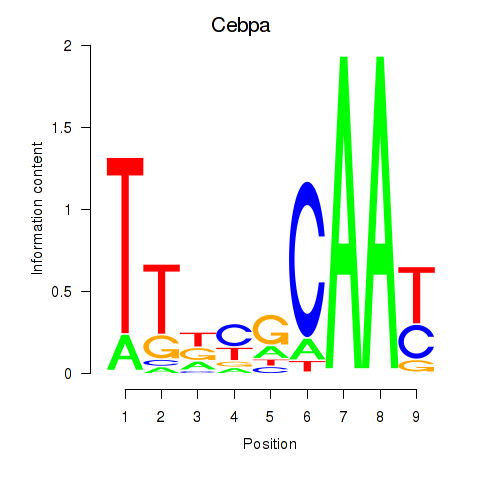
Cebpa
|
ENSRNOG00000010918 | CCAAT/enhancer binding protein alpha |
|
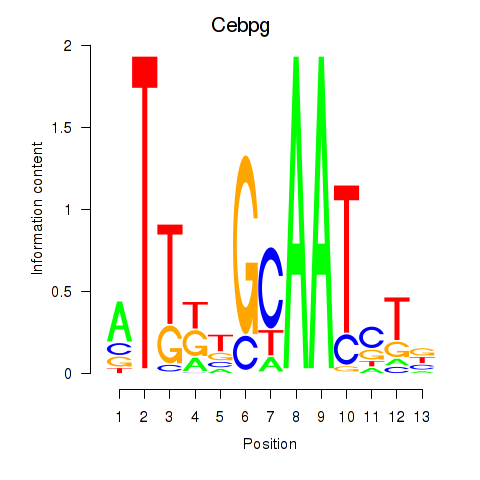
Cebpg
|
ENSRNOG00000021144 | CCAAT/enhancer binding protein gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cebpa | rn6_v1_chr1_+_91363492_91363492 | 0.70 | 2.4e-48 | Click! |
| Cebpg | rn6_v1_chr1_-_91296656_91296656 | 0.16 | 4.3e-03 | Click! |
Activity profile of Cebpa_Cebpg motif
Sorted Z-values of Cebpa_Cebpg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_81717521 | 159.65 |
ENSRNOT00000058422
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr11_-_81444375 | 158.80 |
ENSRNOT00000058479
ENSRNOT00000078131 ENSRNOT00000080949 ENSRNOT00000080562 ENSRNOT00000084867 |
Kng1
|
kininogen 1 |
| chr9_+_9721105 | 132.03 |
ENSRNOT00000073042
ENSRNOT00000075494 |
C3
|
complement C3 |
| chr2_+_104744461 | 106.48 |
ENSRNOT00000016083
ENSRNOT00000082627 |
Cp
|
ceruloplasmin |
| chrX_+_143097525 | 99.60 |
ENSRNOT00000004559
|
F9
|
coagulation factor IX |
| chr10_+_96639924 | 82.75 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chr15_+_57290849 | 82.49 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chr4_+_65110746 | 82.38 |
ENSRNOT00000017675
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr9_+_74124016 | 77.11 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr8_-_111721303 | 70.61 |
ENSRNOT00000045628
ENSRNOT00000012725 |
Tf
|
transferrin |
| chr15_+_28018040 | 63.35 |
ENSRNOT00000041495
|
Rnase4
|
ribonuclease A family member 4 |
| chr14_-_19072677 | 62.90 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr2_+_55775274 | 56.96 |
ENSRNOT00000018545
|
C9
|
complement C9 |
| chr9_-_78969013 | 52.56 |
ENSRNOT00000019772
ENSRNOT00000057585 |
Fn1
|
fibronectin 1 |
| chr19_+_15081158 | 50.47 |
ENSRNOT00000074070
|
Ces1f
|
carboxylesterase 1F |
| chr19_+_15081590 | 50.15 |
ENSRNOT00000024187
|
Ces1f
|
carboxylesterase 1F |
| chr6_+_80188943 | 49.13 |
ENSRNOT00000059335
|
Mia2
|
melanoma inhibitory activity 2 |
| chr9_-_4978892 | 46.88 |
ENSRNOT00000015189
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr11_+_74057361 | 45.35 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr14_+_7113544 | 45.17 |
ENSRNOT00000038188
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr8_+_116857684 | 44.12 |
ENSRNOT00000026711
|
Mst1
|
macrophage stimulating 1 |
| chr1_-_76517134 | 43.70 |
ENSRNOT00000064593
ENSRNOT00000085775 |
LOC100912485
|
alcohol sulfotransferase-like |
| chr7_-_101140308 | 42.61 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr3_-_101474890 | 41.22 |
ENSRNOT00000091869
|
Bbox1
|
gamma-butyrobetaine hydroxylase 1 |
| chr10_-_87407634 | 39.47 |
ENSRNOT00000016657
|
Krt23
|
keratin 23 |
| chr10_+_56710464 | 32.94 |
ENSRNOT00000065370
ENSRNOT00000064064 |
Asgr2
|
asialoglycoprotein receptor 2 |
| chrX_-_110232179 | 32.81 |
ENSRNOT00000014739
|
Serpina7
|
serpin family A member 7 |
| chr1_-_168972725 | 29.22 |
ENSRNOT00000090422
|
Hbb
|
hemoglobin subunit beta |
| chr9_+_46962288 | 27.94 |
ENSRNOT00000082146
|
Il1r1
|
interleukin 1 receptor type 1 |
| chr2_+_158097843 | 27.58 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr4_-_157155609 | 26.80 |
ENSRNOT00000016330
|
C1s
|
complement C1s |
| chr1_-_101596822 | 26.19 |
ENSRNOT00000028490
|
Fgf21
|
fibroblast growth factor 21 |
| chr10_-_62254287 | 25.99 |
ENSRNOT00000004313
|
Serpinf1
|
serpin family F member 1 |
| chr9_+_81656116 | 24.46 |
ENSRNOT00000083421
|
Slc11a1
|
solute carrier family 11 member 1 |
| chr18_-_7081356 | 24.10 |
ENSRNOT00000021253
|
Chst9
|
carbohydrate sulfotransferase 9 |
| chr1_-_22661377 | 22.91 |
ENSRNOT00000021896
|
Vnn3
|
vanin 3 |
| chr2_-_210782746 | 22.76 |
ENSRNOT00000025939
ENSRNOT00000081501 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr8_+_22856539 | 22.66 |
ENSRNOT00000015381
|
Angptl8
|
angiopoietin-like 8 |
| chr4_-_145390447 | 21.14 |
ENSRNOT00000091965
ENSRNOT00000012136 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr3_-_121882726 | 20.71 |
ENSRNOT00000006308
|
Il1b
|
interleukin 1 beta |
| chr4_-_176381477 | 20.68 |
ENSRNOT00000048367
|
Slco1a6
|
solute carrier organic anion transporter family, member 1a6 |
| chr1_+_168964202 | 19.99 |
ENSRNOT00000089102
|
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr9_-_46309451 | 19.55 |
ENSRNOT00000018684
|
Rnf149
|
ring finger protein 149 |
| chr13_+_83073544 | 19.51 |
ENSRNOT00000066119
ENSRNOT00000079796 ENSRNOT00000077070 |
Dpt
|
dermatopontin |
| chrX_+_113510621 | 17.20 |
ENSRNOT00000025912
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr7_-_27240528 | 17.02 |
ENSRNOT00000029435
ENSRNOT00000079731 |
Hsp90b1
|
heat shock protein 90 beta family member 1 |
| chr10_-_82116621 | 16.44 |
ENSRNOT00000003977
ENSRNOT00000051497 ENSRNOT00000085451 |
Abcc3
|
ATP binding cassette subfamily C member 3 |
| chr8_-_7426611 | 16.10 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chrX_+_25016401 | 14.37 |
ENSRNOT00000059270
|
Clcn4
|
chloride voltage-gated channel 4 |
| chr4_+_33890349 | 13.96 |
ENSRNOT00000078680
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr17_-_77527894 | 13.94 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr2_-_235161263 | 13.88 |
ENSRNOT00000080235
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr1_-_78180216 | 13.07 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chrX_+_120624518 | 11.86 |
ENSRNOT00000007967
|
Slc6a14
|
solute carrier family 6 member 14 |
| chr4_+_100166863 | 11.77 |
ENSRNOT00000014505
|
Sftpb
|
surfactant protein B |
| chr3_-_146396299 | 11.54 |
ENSRNOT00000040188
ENSRNOT00000008931 |
Apmap
|
adipocyte plasma membrane associated protein |
| chr5_-_117440254 | 11.52 |
ENSRNOT00000066139
|
Kank4
|
KN motif and ankyrin repeat domains 4 |
| chr8_-_55162421 | 11.30 |
ENSRNOT00000083102
|
Dixdc1
|
DIX domain containing 1 |
| chr14_+_45062662 | 11.17 |
ENSRNOT00000059124
|
Tlr10
|
toll-like receptor 10 |
| chr3_-_11452529 | 10.54 |
ENSRNOT00000020206
|
Slc25a25
|
solute carrier family 25 member 25 |
| chrX_-_154918095 | 10.45 |
ENSRNOT00000085224
|
LOC100911991
|
elongation factor 1-alpha 1-like |
| chr20_-_2103864 | 9.82 |
ENSRNOT00000001014
|
Rnf39
|
ring finger protein 39 |
| chr11_-_69201380 | 9.81 |
ENSRNOT00000085618
|
Mylk
|
myosin light chain kinase |
| chr5_+_151181559 | 9.68 |
ENSRNOT00000085674
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr14_-_14390699 | 9.50 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr6_+_128750795 | 9.48 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr14_-_66978499 | 9.46 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr2_+_60169517 | 9.03 |
ENSRNOT00000080974
|
Prlr
|
prolactin receptor |
| chr14_+_84306466 | 8.90 |
ENSRNOT00000006116
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr18_+_16590197 | 8.87 |
ENSRNOT00000066583
|
Mocos
|
molybdenum cofactor sulfurase |
| chr14_-_8548310 | 8.86 |
ENSRNOT00000092436
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr12_-_20486276 | 8.80 |
ENSRNOT00000074057
|
LOC680910
|
similar to paired immunoglobin-like type 2 receptor beta |
| chr2_-_234296145 | 8.54 |
ENSRNOT00000014155
|
Elovl6
|
ELOVL fatty acid elongase 6 |
| chr12_+_8725517 | 8.52 |
ENSRNOT00000001243
|
Slc46a3
|
solute carrier family 46, member 3 |
| chr12_+_2213705 | 8.23 |
ENSRNOT00000031653
|
Mcemp1
|
mast cell-expressed membrane protein 1 |
| chr1_-_191007503 | 8.23 |
ENSRNOT00000023262
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chrX_+_43881246 | 8.21 |
ENSRNOT00000005153
|
LOC317456
|
hypothetical LOC317456 |
| chr18_-_29562153 | 8.21 |
ENSRNOT00000023977
|
Cd14
|
CD14 molecule |
| chrX_-_153491837 | 7.92 |
ENSRNOT00000089027
|
LOC100360413
|
eukaryotic translation elongation factor 1 alpha 1-like |
| chr2_-_154542919 | 7.88 |
ENSRNOT00000076880
|
Slc33a1
|
solute carrier family 33 member 1 |
| chr10_-_70871066 | 7.87 |
ENSRNOT00000015139
|
Ccl3
|
C-C motif chemokine ligand 3 |
| chr8_+_116754178 | 6.79 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr2_+_212257225 | 6.77 |
ENSRNOT00000077883
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chr5_+_148320438 | 6.57 |
ENSRNOT00000018742
|
Pef1
|
penta-EF hand domain containing 1 |
| chr2_-_154418629 | 6.02 |
ENSRNOT00000076274
ENSRNOT00000076165 |
Plch1
|
phospholipase C, eta 1 |
| chr6_-_42616548 | 5.89 |
ENSRNOT00000081433
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr4_-_23135354 | 5.51 |
ENSRNOT00000011432
|
Steap4
|
STEAP4 metalloreductase |
| chr2_+_58534476 | 5.47 |
ENSRNOT00000077646
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr12_-_21362205 | 5.39 |
ENSRNOT00000064787
|
LOC108348155
|
paired immunoglobulin-like type 2 receptor beta-2 |
| chr13_+_85818427 | 5.15 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr12_+_20667601 | 5.12 |
ENSRNOT00000090081
|
RGD1560281
|
similar to cell surface receptor FDFACT |
| chr1_+_252589785 | 4.47 |
ENSRNOT00000025928
|
Fas
|
Fas cell surface death receptor |
| chr6_-_125723732 | 4.44 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr2_+_234375315 | 4.28 |
ENSRNOT00000071270
|
LOC102549542
|
elongation of very long chain fatty acids protein 6-like |
| chr1_+_168117526 | 4.25 |
ENSRNOT00000051894
|
Olr67
|
olfactory receptor 67 |
| chr9_+_10952374 | 4.22 |
ENSRNOT00000074993
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr16_+_54319377 | 4.15 |
ENSRNOT00000090266
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr2_+_186980793 | 3.94 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr5_+_72103704 | 3.86 |
ENSRNOT00000021629
|
Rad23b
|
RAD23 homolog B, nucleotide excision repair protein |
| chr16_+_61954590 | 3.67 |
ENSRNOT00000017883
|
Rbpms
|
RNA binding protein with multiple splicing |
| chr7_+_2504695 | 3.57 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr1_-_233145924 | 3.48 |
ENSRNOT00000018763
ENSRNOT00000082442 |
Psat1
|
phosphoserine aminotransferase 1 |
| chr7_-_123572082 | 3.04 |
ENSRNOT00000068020
|
Naga
|
N-acetyl galactosaminidase, alpha |
| chr13_-_73390393 | 2.88 |
ENSRNOT00000067253
ENSRNOT00000093438 |
Lhx4
|
LIM homeobox 4 |
| chr1_-_80809769 | 2.81 |
ENSRNOT00000045073
|
Ceacam19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr8_+_2604962 | 2.57 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chr4_+_1417537 | 2.52 |
ENSRNOT00000072547
|
Olr1230
|
olfactory receptor 1230 |
| chr20_-_848229 | 2.52 |
ENSRNOT00000000974
|
Olr1694
|
olfactory receptor 1694 |
| chr1_+_222861777 | 2.47 |
ENSRNOT00000090872
|
Pla2g16
|
phospholipase A2, group XVI |
| chr16_+_61954809 | 2.46 |
ENSRNOT00000068011
|
Rbpms
|
RNA binding protein with multiple splicing |
| chr7_+_16461899 | 2.26 |
ENSRNOT00000051359
|
Olr1055
|
olfactory receptor 1055 |
| chr7_-_9174330 | 2.18 |
ENSRNOT00000051063
|
Olr1060
|
olfactory receptor 1060 |
| chr4_+_1658278 | 2.15 |
ENSRNOT00000073845
|
Olr1250
|
olfactory receptor 1250 |
| chr17_+_39032661 | 2.15 |
ENSRNOT00000022381
|
Prl7a3
|
prolactin family 7, subfamily a, member 3 |
| chr1_-_228524439 | 2.12 |
ENSRNOT00000028601
|
Olr323
|
olfactory receptor 323 |
| chr3_-_102672294 | 1.94 |
ENSRNOT00000030909
|
Olr760
|
olfactory receptor 760 |
| chr1_+_162320730 | 1.88 |
ENSRNOT00000035743
|
Kctd21
|
potassium channel tetramerization domain containing 21 |
| chr2_+_186980992 | 1.88 |
ENSRNOT00000020717
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr3_+_73064092 | 1.83 |
ENSRNOT00000072178
|
LOC684539
|
similar to olfactory receptor Olr490 |
| chr14_+_17492081 | 1.78 |
ENSRNOT00000003346
|
G3bp2
|
G3BP stress granule assembly factor 2 |
| chr4_-_170129407 | 1.76 |
ENSRNOT00000075717
|
LOC100912609
|
uncharacterized LOC100912609 |
| chr18_+_35249137 | 1.75 |
ENSRNOT00000017489
|
LOC100910942
|
serine protease inhibitor Kazal-type 6-like |
| chr8_-_40192831 | 1.71 |
ENSRNOT00000048448
|
Olr1194
|
olfactory receptor 1194 |
| chr11_+_55497234 | 1.70 |
ENSRNOT00000070882
|
LOC108348268
|
olfactory receptor 2T33-like |
| chr13_+_52887649 | 1.70 |
ENSRNOT00000048033
|
Ascl5
|
achaete-scute family bHLH transcription factor 5 |
| chr7_-_7311272 | 1.66 |
ENSRNOT00000040812
|
Olr1020
|
olfactory receptor 1020 |
| chr5_-_59913348 | 1.64 |
ENSRNOT00000018164
|
LOC100359916
|
heterogeneous nuclear ribonucleoprotein K-like |
| chrX_-_75224268 | 1.62 |
ENSRNOT00000089809
|
Abcb7
|
ATP binding cassette subfamily B member 7 |
| chr2_+_58448917 | 1.62 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr14_+_900696 | 1.62 |
ENSRNOT00000049544
|
Vom2r71
|
vomeronasal 2 receptor, 71 |
| chr4_+_84478839 | 1.56 |
ENSRNOT00000012668
|
Prr15
|
proline rich 15 |
| chr3_+_16817051 | 1.53 |
ENSRNOT00000071666
|
AABR07051550.1
|
|
| chr10_-_83898527 | 1.47 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr2_-_1561464 | 1.25 |
ENSRNOT00000062055
ENSRNOT00000062054 ENSRNOT00000062052 |
Cast
|
calpastatin |
| chr3_-_103140972 | 1.12 |
ENSRNOT00000052368
|
Olr780
|
olfactory receptor 780 |
| chrX_-_63809861 | 1.05 |
ENSRNOT00000009870
|
Maged1
|
MAGE family member D1 |
| chr12_-_20660304 | 1.02 |
ENSRNOT00000091323
|
RGD1561730
|
similar to cell surface receptor FDFACT |
| chr7_-_9136515 | 0.96 |
ENSRNOT00000081964
|
Olr1059
|
olfactory receptor 1059 |
| chr2_+_94266213 | 0.96 |
ENSRNOT00000090023
|
Pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr1_+_252323865 | 0.95 |
ENSRNOT00000026299
|
Lipk
|
lipase, family member K |
| chr3_-_160573978 | 0.85 |
ENSRNOT00000018386
|
Wfdc5
|
WAP four-disulfide core domain 5 |
| chr12_+_44046454 | 0.80 |
ENSRNOT00000064756
|
Fbxw8
|
F-box and WD repeat domain containing 8 |
| chr15_-_46166335 | 0.63 |
ENSRNOT00000059215
|
Defb42
|
defensin beta 42 |
| chr7_-_9104719 | 0.54 |
ENSRNOT00000088247
|
LOC103692758
|
olfactory receptor 6C74-like |
| chr11_+_43431431 | 0.53 |
ENSRNOT00000081261
|
Olr1545
|
olfactory receptor 1545 |
| chr10_+_107502695 | 0.49 |
ENSRNOT00000038088
|
Engase
|
endo-beta-N-acetylglucosaminidase |
| chr1_+_91152635 | 0.49 |
ENSRNOT00000073438
|
LOC100912070
|
dermokine-like |
| chr7_-_75569778 | 0.48 |
ENSRNOT00000040687
|
LOC362901
|
Ac1254 |
| chr3_-_74701712 | 0.41 |
ENSRNOT00000041127
|
Olr541
|
olfactory receptor 541 |
| chr20_-_10013559 | 0.40 |
ENSRNOT00000091623
|
Rsph1
|
radial spoke head 1 homolog |
| chr10_+_61033113 | 0.34 |
ENSRNOT00000043016
|
Olr1513
|
olfactory receptor 1513 |
| chr2_-_58534211 | 0.33 |
ENSRNOT00000089178
|
Skp2
|
S-phase kinase associated protein 2 |
| chr15_+_41643541 | 0.32 |
ENSRNOT00000019646
|
Arl11
|
ADP-ribosylation factor like GTPase 11 |
| chr1_-_82279145 | 0.31 |
ENSRNOT00000057433
|
Cxcl17
|
C-X-C motif chemokine ligand 17 |
| chr1_-_74638587 | 0.31 |
ENSRNOT00000046047
|
Vom2r30
|
vomeronasal 2 receptor, 30 |
| chr1_+_112976770 | 0.26 |
ENSRNOT00000079348
|
Gabrb3
|
gamma-aminobutyric acid type A receptor beta 3 subunit |
| chr7_+_23854846 | 0.25 |
ENSRNOT00000037290
|
Bpifc
|
BPI fold containing family C |
| chr3_+_73366155 | 0.25 |
ENSRNOT00000044979
|
Olr473
|
olfactory receptor 473 |
| chr3_-_14386118 | 0.23 |
ENSRNOT00000025649
|
Rab14
|
RAB14, member RAS oncogene family |
| chr1_+_67917477 | 0.13 |
ENSRNOT00000052217
|
Vom1r40
|
vomeronasal 1 receptor 40 |
| chr18_-_60002529 | 0.05 |
ENSRNOT00000081014
ENSRNOT00000024264 ENSRNOT00000059162 |
Nars
|
asparaginyl-tRNA synthetase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cebpa_Cebpg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 44.0 | 132.0 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 27.5 | 82.5 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 20.7 | 82.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 17.7 | 70.6 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 17.5 | 52.6 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 15.4 | 77.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 13.3 | 106.5 | GO:0015679 | plasma membrane copper ion transport(GO:0015679) |
| 9.3 | 27.9 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 8.2 | 49.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 8.2 | 24.5 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 7.5 | 22.5 | GO:0071623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 7.3 | 29.2 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 6.9 | 159.6 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 6.9 | 20.7 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 6.9 | 27.6 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 6.9 | 41.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 6.3 | 82.4 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 6.3 | 44.1 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 5.7 | 57.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 5.4 | 16.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 5.2 | 26.2 | GO:1904640 | response to methionine(GO:1904640) |
| 5.1 | 30.5 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 4.7 | 14.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 4.0 | 158.8 | GO:0042311 | vasodilation(GO:0042311) |
| 3.2 | 9.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 3.0 | 32.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 2.7 | 16.4 | GO:0042908 | canalicular bile acid transport(GO:0015722) xenobiotic transport(GO:0042908) |
| 2.5 | 9.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 2.4 | 11.8 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 1.9 | 26.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.8 | 12.8 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 1.6 | 11.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 1.5 | 22.7 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 1.5 | 21.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 1.4 | 20.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.4 | 6.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.3 | 7.9 | GO:0051182 | coenzyme transport(GO:0051182) |
| 1.3 | 7.9 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.3 | 3.9 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 1.2 | 10.5 | GO:0015866 | ADP transport(GO:0015866) |
| 1.1 | 3.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 1.0 | 9.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.9 | 5.5 | GO:0015677 | copper ion import(GO:0015677) |
| 0.9 | 8.2 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.9 | 17.0 | GO:0071287 | cellular response to manganese ion(GO:0071287) |
| 0.9 | 3.6 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.9 | 8.9 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.8 | 8.9 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.8 | 20.7 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.7 | 2.9 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.7 | 99.0 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.7 | 3.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.7 | 22.9 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.6 | 2.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.6 | 4.4 | GO:0048251 | elastic fiber assembly(GO:0048251) regulation of removal of superoxide radicals(GO:2000121) |
| 0.5 | 9.7 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.5 | 3.0 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.5 | 19.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.4 | 6.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.4 | 25.2 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.4 | 63.3 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.4 | 30.3 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.3 | 14.4 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.3 | 2.5 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.3 | 18.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.3 | 5.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.2 | 7.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.2 | 4.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 55.1 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.2 | 6.8 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.2 | 4.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 1.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 1.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 1.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 11.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 9.8 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 9.5 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 8.9 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.0 | 0.8 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 1.6 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 1.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.0 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 2.2 | GO:0007565 | female pregnancy(GO:0007565) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.2 | 57.0 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 11.8 | 70.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 8.2 | 24.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 7.3 | 29.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 6.6 | 52.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 6.4 | 82.8 | GO:0042627 | chylomicron(GO:0042627) |
| 5.7 | 629.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 3.2 | 26.0 | GO:0043203 | axon hillock(GO:0043203) |
| 2.9 | 20.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 2.6 | 49.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 1.6 | 8.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.5 | 17.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.4 | 97.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 1.4 | 71.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 1.3 | 3.9 | GO:0071942 | XPC complex(GO:0071942) |
| 1.2 | 5.9 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 1.1 | 4.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.0 | 12.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.9 | 66.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.9 | 4.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.6 | 9.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.5 | 6.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.5 | 2.6 | GO:0072558 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.3 | 488.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.3 | 13.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 39.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.3 | 6.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 9.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 9.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 14.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.2 | 11.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.8 | GO:1990393 | Cul7-RING ubiquitin ligase complex(GO:0031467) 3M complex(GO:1990393) |
| 0.1 | 1.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 8.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 19.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 18.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 60.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 4.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 6.7 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 44.0 | 132.0 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 39.9 | 159.6 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 27.6 | 82.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 27.5 | 82.4 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 17.7 | 106.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 11.7 | 46.9 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 9.7 | 29.2 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 9.3 | 27.9 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 8.8 | 70.6 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 8.2 | 49.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 7.5 | 52.6 | GO:0045340 | mercury ion binding(GO:0045340) |
| 6.4 | 77.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 5.0 | 44.6 | GO:0046790 | virion binding(GO:0046790) |
| 4.7 | 14.0 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 4.4 | 13.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 4.1 | 32.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 4.0 | 24.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 3.1 | 82.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 3.0 | 9.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 2.6 | 7.9 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 2.5 | 9.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 2.4 | 24.5 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 2.4 | 9.5 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 2.2 | 8.9 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 2.1 | 148.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 2.0 | 6.0 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 2.0 | 20.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.9 | 9.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.8 | 16.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 1.6 | 11.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.6 | 8.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 1.6 | 9.7 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.6 | 7.9 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 1.5 | 6.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 1.5 | 10.5 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 1.3 | 12.8 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.1 | 20.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.9 | 5.5 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.9 | 22.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.9 | 4.5 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.9 | 5.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.8 | 18.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.8 | 41.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.8 | 26.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.8 | 14.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.7 | 165.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.7 | 5.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.6 | 6.8 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.6 | 22.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.6 | 92.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.5 | 53.7 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.4 | 11.9 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.4 | 9.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.3 | 11.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 6.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 3.0 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.3 | 6.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 18.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.2 | 22.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 2.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 3.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 18.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 4.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 18.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 3.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 11.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 7.9 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 9.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 290.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 3.5 | 52.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 2.9 | 154.1 | PID BMP PATHWAY | BMP receptor signaling |
| 2.7 | 70.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.7 | 23.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 1.4 | 101.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 1.3 | 53.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 1.1 | 27.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.9 | 27.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.8 | 175.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.5 | 8.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 4.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 6.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.4 | 9.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.4 | 24.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.4 | 17.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.3 | 15.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.3 | 9.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 5.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 7.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 11.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 18.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 24.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.3 | 258.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 9.4 | 132.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 6.9 | 82.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 4.5 | 130.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 4.0 | 83.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 3.1 | 52.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 2.6 | 76.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 1.5 | 17.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 1.5 | 16.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.4 | 8.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 1.0 | 48.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 1.0 | 24.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.6 | 9.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.5 | 11.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 107.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.5 | 8.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.5 | 9.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.4 | 9.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 9.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 4.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 5.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.3 | 14.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 7.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 6.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 5.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 2.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 3.9 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 2.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 7.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 5.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 8.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 6.3 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.1 | 1.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 6.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |