Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
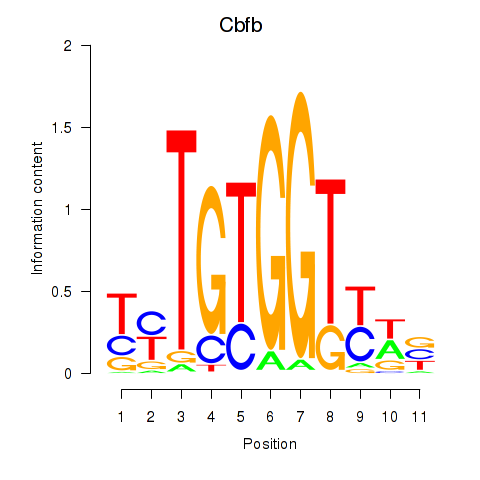
Results for Cbfb
Z-value: 0.58
Transcription factors associated with Cbfb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cbfb
|
ENSRNOG00000014647 | core-binding factor, beta subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cbfb | rn6_v1_chr19_+_37127508_37127508 | 0.51 | 9.1e-23 | Click! |
Activity profile of Cbfb motif
Sorted Z-values of Cbfb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_88763733 | 29.10 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr3_-_44177689 | 26.18 |
ENSRNOT00000006387
|
Cytip
|
cytohesin 1 interacting protein |
| chr2_-_88553086 | 24.27 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr2_-_88660449 | 24.16 |
ENSRNOT00000051741
|
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr14_+_3058993 | 16.06 |
ENSRNOT00000002807
|
Gfi1
|
growth factor independent 1 transcriptional repressor |
| chr11_-_33003021 | 13.73 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr1_+_229642412 | 13.22 |
ENSRNOT00000017109
|
Lpxn
|
leupaxin |
| chr3_+_11756384 | 13.18 |
ENSRNOT00000087762
|
Sh2d3c
|
SH2 domain containing 3C |
| chr2_+_206342066 | 12.91 |
ENSRNOT00000026556
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr14_-_100184192 | 10.32 |
ENSRNOT00000007044
|
Plek
|
pleckstrin |
| chr10_-_70871066 | 10.04 |
ENSRNOT00000015139
|
Ccl3
|
C-C motif chemokine ligand 3 |
| chr4_-_170740274 | 9.02 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr12_+_9360672 | 7.31 |
ENSRNOT00000088957
|
Flt3
|
fms-related tyrosine kinase 3 |
| chr15_+_86243148 | 6.66 |
ENSRNOT00000084471
ENSRNOT00000090727 |
Lmo7
|
LIM domain 7 |
| chr19_+_25239674 | 5.26 |
ENSRNOT00000037453
|
Podnl1
|
podocan-like 1 |
| chr3_+_170399302 | 5.05 |
ENSRNOT00000035245
|
Cass4
|
Cas scaffolding protein family member 4 |
| chr14_-_6679878 | 4.14 |
ENSRNOT00000075989
ENSRNOT00000067875 |
Spp1
|
secreted phosphoprotein 1 |
| chr10_-_39604070 | 2.91 |
ENSRNOT00000032333
|
Csf2
|
colony stimulating factor 2 |
| chr9_+_60039297 | 2.47 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr18_+_29191731 | 1.88 |
ENSRNOT00000068085
ENSRNOT00000025495 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr5_+_101526551 | 1.80 |
ENSRNOT00000015254
|
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chrX_+_120624518 | 1.72 |
ENSRNOT00000007967
|
Slc6a14
|
solute carrier family 6 member 14 |
| chr7_+_61337381 | 1.59 |
ENSRNOT00000009919
|
Ifng
|
interferon gamma |
| chr10_+_104320743 | 1.21 |
ENSRNOT00000005684
|
Tmem94
|
transmembrane protein 94 |
| chr4_-_176922424 | 0.91 |
ENSRNOT00000055540
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr4_-_7342410 | 0.84 |
ENSRNOT00000013058
|
Nos3
|
nitric oxide synthase 3 |
| chr1_+_167522895 | 0.52 |
ENSRNOT00000092987
|
Stim1
|
stromal interaction molecule 1 |
| chr5_-_137928107 | 0.24 |
ENSRNOT00000048274
|
Olr869
|
olfactory receptor 869 |
| chr2_-_252505971 | 0.17 |
ENSRNOT00000021984
|
LOC56764
|
dnaj-like protein |
| chr18_+_17550350 | 0.06 |
ENSRNOT00000078870
|
RGD1562608
|
similar to KIAA1328 protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cbfb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.1 | GO:0070103 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 5.1 | 20.2 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 4.6 | 13.7 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 2.6 | 10.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.9 | 77.5 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 1.7 | 10.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.7 | 13.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 1.4 | 4.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.6 | 2.9 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 2.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 9.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.2 | 0.9 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.1 | 5.3 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 10.4 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 32.8 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 13.2 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 0.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 16.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 13.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 10.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 26.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 12.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 6.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 81.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 7.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 4.1 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 5.3 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 10.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 1.0 | 77.5 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.5 | 13.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.5 | 9.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 10.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 13.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 0.9 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 7.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 0.8 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 12.9 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 6.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 16.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 2.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 4.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 5.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 13.2 | GO:0003712 | transcription cofactor activity(GO:0003712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 13.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 16.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 11.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 5.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 10.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 4.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |