Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Bsx
Z-value: 0.51
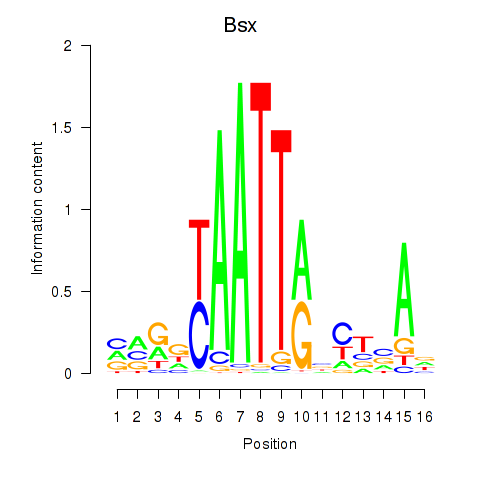
Transcription factors associated with Bsx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bsx
|
ENSRNOG00000024438 | brain specific homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bsx | rn6_v1_chr8_+_45061670_45061670 | 0.26 | 1.6e-06 | Click! |
Activity profile of Bsx motif
Sorted Z-values of Bsx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_133959447 | 18.37 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr9_-_85243001 | 18.14 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr12_+_47179664 | 17.22 |
ENSRNOT00000001551
|
Cabp1
|
calcium binding protein 1 |
| chr7_-_76488216 | 16.73 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr7_-_76294663 | 16.63 |
ENSRNOT00000064513
|
Ncald
|
neurocalcin delta |
| chr5_-_130085838 | 16.51 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr9_+_73418607 | 15.83 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr8_-_82533689 | 15.45 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr6_+_58468155 | 13.21 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr14_+_3882398 | 12.83 |
ENSRNOT00000033794
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog |
| chr12_-_19167015 | 12.80 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr15_+_56666012 | 12.59 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr15_+_11298478 | 11.47 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chrX_+_6273733 | 11.09 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr19_+_6046665 | 10.85 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chr2_-_219262901 | 10.61 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr1_+_48077033 | 10.45 |
ENSRNOT00000020100
|
Mas1
|
MAS1 proto-oncogene, G protein-coupled receptor |
| chr8_+_33239139 | 10.17 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr5_+_43603043 | 9.82 |
ENSRNOT00000009899
|
Epha7
|
Eph receptor A7 |
| chr18_-_24735349 | 9.81 |
ENSRNOT00000036537
|
Gpr17
|
G protein-coupled receptor 17 |
| chrX_-_10031167 | 9.76 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chr8_+_82038967 | 9.25 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr1_-_67065797 | 9.12 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr1_+_28454966 | 8.55 |
ENSRNOT00000078841
ENSRNOT00000030327 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr17_-_87826421 | 8.47 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr18_+_51523758 | 7.81 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr1_+_265157379 | 6.67 |
ENSRNOT00000022426
|
Btrc
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr1_+_273854248 | 6.22 |
ENSRNOT00000044827
ENSRNOT00000017600 ENSRNOT00000086496 |
Add3
|
adducin 3 |
| chr9_-_44419998 | 6.06 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr4_-_61720956 | 5.97 |
ENSRNOT00000012879
|
Akr1b1
|
aldo-keto reductase family 1 member B |
| chr5_+_154800226 | 4.87 |
ENSRNOT00000016046
|
Htr1d
|
5-hydroxytryptamine receptor 1D |
| chr2_-_113616766 | 4.85 |
ENSRNOT00000016858
ENSRNOT00000074723 |
Tmem212
|
transmembrane protein 212 |
| chr16_+_22979444 | 4.75 |
ENSRNOT00000017822
|
Csgalnact1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr5_+_137257287 | 4.63 |
ENSRNOT00000037160
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr2_+_145174876 | 4.63 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr1_+_217345545 | 4.35 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr8_+_119524039 | 4.31 |
ENSRNOT00000056050
|
Epm2aip1
|
EPM2A interacting protein 1 |
| chr4_+_56981283 | 4.20 |
ENSRNOT00000010989
|
Tspan33
|
tetraspanin 33 |
| chr4_-_168656673 | 4.00 |
ENSRNOT00000009341
|
Gpr19
|
G protein-coupled receptor 19 |
| chr11_-_90234286 | 3.39 |
ENSRNOT00000071986
|
Efcab1
|
EF hand calcium binding domain 1 |
| chr7_-_140502441 | 3.27 |
ENSRNOT00000089544
|
Kmt2d
|
lysine methyltransferase 2D |
| chrX_+_9956370 | 3.00 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr3_-_101547478 | 2.82 |
ENSRNOT00000006203
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr11_-_70585053 | 2.80 |
ENSRNOT00000086955
|
Zfp148
|
zinc finger protein 148 |
| chrX_-_84768463 | 2.55 |
ENSRNOT00000088570
|
Chm
|
CHM, Rab escort protein 1 |
| chr4_-_67301102 | 2.49 |
ENSRNOT00000034549
|
Dennd2a
|
DENN domain containing 2A |
| chr18_-_69780922 | 2.35 |
ENSRNOT00000079093
|
Me2
|
malic enzyme 2 |
| chr10_+_109107389 | 2.33 |
ENSRNOT00000068437
ENSRNOT00000005687 |
Baiap2
|
BAI1-associated protein 2 |
| chr1_+_80279706 | 2.31 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr4_+_87312766 | 2.24 |
ENSRNOT00000052126
|
Vom1r71
|
vomeronasal 1 receptor 71 |
| chr2_-_149432106 | 2.21 |
ENSRNOT00000051027
|
P2ry13
|
purinergic receptor P2Y13 |
| chr12_+_22641104 | 2.08 |
ENSRNOT00000001916
|
Serpine1
|
serpin family E member 1 |
| chr2_-_149088787 | 1.99 |
ENSRNOT00000064833
|
Clrn1
|
clarin 1 |
| chr2_-_227582895 | 1.85 |
ENSRNOT00000020574
|
Mettl14
|
methyltransferase like 14 |
| chr4_+_32541706 | 1.81 |
ENSRNOT00000050827
|
Sdhaf3
|
succinate dehydrogenase complex assembly factor 3 |
| chr8_+_70447040 | 1.56 |
ENSRNOT00000016798
|
Ints14
|
integrator complex subunit 14 |
| chr3_+_148654668 | 1.55 |
ENSRNOT00000081370
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr10_+_44228800 | 1.49 |
ENSRNOT00000046817
|
Olr1431
|
olfactory receptor 1431 |
| chr1_-_277902279 | 1.34 |
ENSRNOT00000078416
|
Ablim1
|
actin-binding LIM protein 1 |
| chr1_+_213597478 | 1.33 |
ENSRNOT00000017562
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr8_+_42663633 | 1.24 |
ENSRNOT00000048537
|
Olr1266
|
olfactory receptor 1266 |
| chr1_-_64147251 | 1.18 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr3_-_76102782 | 1.18 |
ENSRNOT00000007690
|
Olr602
|
olfactory receptor 602 |
| chr20_+_1172867 | 1.09 |
ENSRNOT00000041189
|
Olr1708
|
olfactory receptor 1708 |
| chr18_-_26656879 | 1.01 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chrX_-_70835089 | 0.93 |
ENSRNOT00000076351
|
Tex11
|
testis expressed 11 |
| chr1_+_225037737 | 0.92 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr13_-_89545182 | 0.90 |
ENSRNOT00000078402
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr5_+_137564607 | 0.83 |
ENSRNOT00000075742
|
LOC100912529
|
olfactory receptor 2B6-like |
| chr6_+_124123228 | 0.82 |
ENSRNOT00000005329
|
Psmc1
|
proteasome 26S subunit, ATPase 1 |
| chr5_-_164648328 | 0.76 |
ENSRNOT00000090811
|
Zfp933
|
zinc finger protein 933 |
| chr18_-_48720472 | 0.58 |
ENSRNOT00000023236
|
Cep120
|
centrosomal protein 120 |
| chr1_-_170694872 | 0.58 |
ENSRNOT00000075443
|
LOC100911839
|
olfactory receptor 2D3-like |
| chr17_-_42740021 | 0.56 |
ENSRNOT00000023063
|
Prl3a1
|
Prolactin family 3, subfamily a, member 1 |
| chr9_-_98551410 | 0.51 |
ENSRNOT00000066346
ENSRNOT00000086678 |
Hes6
|
hes family bHLH transcription factor 6 |
| chr7_+_18068060 | 0.48 |
ENSRNOT00000065474
|
Vom1r107
|
vomeronasal 1 receptor 107 |
| chr7_+_23854846 | 0.47 |
ENSRNOT00000037290
|
Bpifc
|
BPI fold containing family C |
| chr4_+_87516218 | 0.45 |
ENSRNOT00000090182
|
Vom1r66
|
vomeronasal 1 receptor 66 |
| chr1_+_219833299 | 0.29 |
ENSRNOT00000087432
|
Pc
|
pyruvate carboxylase |
| chr13_+_67545430 | 0.27 |
ENSRNOT00000003405
|
Pdc
|
phosducin |
| chr16_+_26593370 | 0.20 |
ENSRNOT00000065453
|
AABR07025051.1
|
|
| chr3_+_102529184 | 0.20 |
ENSRNOT00000040444
|
Olr752
|
olfactory receptor 752 |
| chr3_+_77047466 | 0.14 |
ENSRNOT00000089424
|
Olr648
|
olfactory receptor 648 |
| chr1_+_255579474 | 0.10 |
ENSRNOT00000024465
|
Btaf1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr1_-_140835151 | 0.08 |
ENSRNOT00000032174
|
Hapln3
|
hyaluronan and proteoglycan link protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Bsx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 2.3 | 9.3 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 2.1 | 10.6 | GO:0061743 | motor learning(GO:0061743) |
| 2.0 | 6.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.9 | 15.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.9 | 7.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 1.8 | 11.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.6 | 9.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 1.6 | 4.8 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 1.4 | 17.2 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 1.2 | 18.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 1.1 | 13.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.7 | 10.4 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.7 | 2.1 | GO:1904638 | response to resveratrol(GO:1904638) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.7 | 4.6 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.7 | 3.3 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.6 | 4.9 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.5 | 9.8 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.5 | 4.3 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.4 | 7.9 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.4 | 15.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.4 | 2.3 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.4 | 1.8 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.3 | 4.3 | GO:0099562 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.3 | 2.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 31.1 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.3 | 6.7 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.2 | 2.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 2.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 2.3 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.2 | 18.4 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.2 | 10.9 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 2.3 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 1.5 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 12.8 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.6 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.3 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 8.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.1 | 2.8 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 4.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 2.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 9.5 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 4.2 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 7.4 | GO:0030031 | cell projection assembly(GO:0030031) |
| 0.0 | 2.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.8 | 15.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.7 | 16.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.2 | 6.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.1 | 18.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.8 | 12.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.7 | 10.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.6 | 2.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.6 | 12.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.5 | 4.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 1.9 | GO:0036396 | MIS complex(GO:0036396) |
| 0.3 | 4.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 3.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 2.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 7.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 6.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 15.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 6.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 27.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 1.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 2.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 4.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 21.1 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 5.0 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 1.6 | 4.8 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 1.5 | 10.4 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 1.3 | 17.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 1.2 | 33.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 1.0 | 9.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.9 | 15.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.7 | 15.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.6 | 18.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.6 | 1.9 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.6 | 16.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.5 | 9.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.5 | 6.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.5 | 2.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 4.6 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 18.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 4.9 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.4 | 2.3 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 4.3 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.3 | 11.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 2.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 12.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.2 | 2.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 12.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 6.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 4.7 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 10.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 18.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 13.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 2.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 8.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 13.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 15.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 9.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 10.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 6.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 33.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.2 | 17.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.0 | 12.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.4 | 10.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 9.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 4.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 4.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 8.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 6.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.8 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |