Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
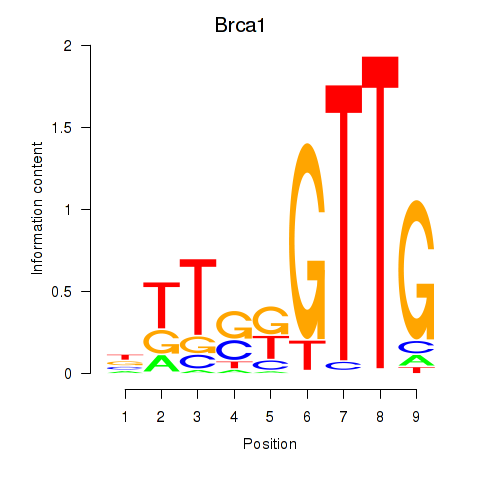
Results for Brca1
Z-value: 0.48
Transcription factors associated with Brca1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Brca1
|
ENSRNOG00000020701 | BRCA1, DNA repair associated |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Brca1 | rn6_v1_chr10_-_89454681_89454681 | -0.09 | 1.1e-01 | Click! |
Activity profile of Brca1 motif
Sorted Z-values of Brca1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_47397558 | 13.90 |
ENSRNOT00000085923
|
Epdr1
|
ependymin related 1 |
| chr1_-_143392532 | 12.55 |
ENSRNOT00000026089
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr3_-_48535909 | 9.49 |
ENSRNOT00000008148
|
Fap
|
fibroblast activation protein, alpha |
| chr13_-_91872954 | 8.77 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr4_+_154215250 | 7.76 |
ENSRNOT00000072465
|
Mug2
|
murinoglobulin 2 |
| chr3_+_14467330 | 7.53 |
ENSRNOT00000078939
|
Gsn
|
gelsolin |
| chr14_-_19159923 | 7.18 |
ENSRNOT00000003879
|
Afp
|
alpha-fetoprotein |
| chr5_+_152533349 | 7.11 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr20_+_31102476 | 6.48 |
ENSRNOT00000078719
|
Lrrc20
|
leucine rich repeat containing 20 |
| chr8_-_49502647 | 5.08 |
ENSRNOT00000040313
|
Tmprss4
|
transmembrane protease, serine 4 |
| chrX_-_23139694 | 4.77 |
ENSRNOT00000033656
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr10_-_51669297 | 4.65 |
ENSRNOT00000071595
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr13_-_83680207 | 4.44 |
ENSRNOT00000004199
|
Dcaf6
|
DDB1 and CUL4 associated factor 6 |
| chr16_-_75637789 | 3.83 |
ENSRNOT00000058029
|
Defb4
|
defensin beta 4 |
| chr3_-_101474890 | 3.73 |
ENSRNOT00000091869
|
Bbox1
|
gamma-butyrobetaine hydroxylase 1 |
| chr18_+_53088994 | 3.33 |
ENSRNOT00000091921
|
AC104053.2
|
|
| chr3_+_100787956 | 3.19 |
ENSRNOT00000073636
|
Bdnf
|
brain-derived neurotrophic factor |
| chr10_+_18912865 | 3.15 |
ENSRNOT00000007393
|
Kcnmb1
|
potassium calcium-activated channel subfamily M regulatory beta subunit 1 |
| chr7_-_143167772 | 3.06 |
ENSRNOT00000011374
|
Krt85
|
keratin 85 |
| chr14_-_86190659 | 2.94 |
ENSRNOT00000086474
|
Gck
|
glucokinase |
| chr10_-_89816491 | 2.93 |
ENSRNOT00000028220
|
Meox1
|
mesenchyme homeobox 1 |
| chr2_-_147819335 | 2.61 |
ENSRNOT00000057909
|
Ankub1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr1_-_94610854 | 2.60 |
ENSRNOT00000020383
|
Plekhf1
|
pleckstrin homology and FYVE domain containing 1 |
| chr16_+_74408183 | 2.60 |
ENSRNOT00000036506
|
Smim19
|
small integral membrane protein 19 |
| chr10_-_90042877 | 2.54 |
ENSRNOT00000028318
|
Ppy
|
pancreatic polypeptide |
| chr3_-_72895740 | 2.51 |
ENSRNOT00000012568
|
Fads2l1
|
fatty acid desaturase 2-like 1 |
| chr16_+_54386513 | 2.49 |
ENSRNOT00000080696
|
AABR07025896.1
|
|
| chr15_-_8183682 | 2.43 |
ENSRNOT00000011438
|
Nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr11_-_15858281 | 2.43 |
ENSRNOT00000090586
|
AABR07033287.1
|
|
| chr8_+_26432514 | 2.36 |
ENSRNOT00000081574
|
Sept7
|
septin 7 |
| chr11_-_87485833 | 2.29 |
ENSRNOT00000079948
|
AABR07034751.1
|
|
| chr15_-_52210746 | 2.21 |
ENSRNOT00000046054
|
Bmp1
|
bone morphogenetic protein 1 |
| chr5_+_104362971 | 2.13 |
ENSRNOT00000058520
|
Adamtsl1
|
ADAMTS-like 1 |
| chr14_-_92577936 | 2.06 |
ENSRNOT00000086154
|
Cobl
|
cordon-bleu WH2 repeat protein |
| chr6_+_28515025 | 2.05 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr19_-_55183557 | 2.03 |
ENSRNOT00000017317
|
Mlnr
|
motilin receptor |
| chr2_+_93803449 | 2.01 |
ENSRNOT00000014949
|
Fabp9
|
fatty acid binding protein 9 |
| chr10_+_66099531 | 1.99 |
ENSRNOT00000056192
|
Lyrm9
|
LYR motif containing 9 |
| chr12_-_20028097 | 1.96 |
ENSRNOT00000048430
|
Vom2r62
|
vomeronasal 2 receptor, 62 |
| chr9_+_53626592 | 1.96 |
ENSRNOT00000031267
|
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr1_+_264309214 | 1.95 |
ENSRNOT00000019070
|
Hif1an
|
hypoxia-inducible factor 1, alpha subunit inhibitor |
| chrX_+_45637415 | 1.95 |
ENSRNOT00000050544
|
RGD1563378
|
similar to ferritin heavy polypeptide-like 17 |
| chr3_-_139735397 | 1.90 |
ENSRNOT00000088315
|
AABR07054088.1
|
|
| chr1_+_214747625 | 1.86 |
ENSRNOT00000091110
|
AABR07006033.1
|
|
| chr13_+_106751625 | 1.85 |
ENSRNOT00000004992
|
Ush2a
|
usherin |
| chr10_+_89251370 | 1.82 |
ENSRNOT00000076820
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr19_-_14945302 | 1.76 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr17_+_36690249 | 1.71 |
ENSRNOT00000089476
ENSRNOT00000082140 |
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chr12_+_10255416 | 1.68 |
ENSRNOT00000092720
|
Gpr12
|
G protein-coupled receptor 12 |
| chr20_-_47306318 | 1.66 |
ENSRNOT00000075151
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr2_-_243407608 | 1.63 |
ENSRNOT00000014631
|
Mttp
|
microsomal triglyceride transfer protein |
| chr11_-_32550539 | 1.62 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr9_+_51009116 | 1.59 |
ENSRNOT00000039313
|
Mettl21cl1
|
methyltransferase like 21C-like 1 |
| chr13_+_63526486 | 1.58 |
ENSRNOT00000003788
|
Brinp3
|
BMP/retinoic acid inducible neural specific 3 |
| chr16_+_69022792 | 1.54 |
ENSRNOT00000035985
|
Got1l1
|
glutamic-oxaloacetic transaminase 1-like 1 |
| chr13_-_102790639 | 1.51 |
ENSRNOT00000003177
|
RGD1310587
|
similar to hypothetical protein FLJ14146 |
| chr13_+_98857177 | 1.51 |
ENSRNOT00000004232
|
Parp1
|
poly (ADP-ribose) polymerase 1 |
| chr3_-_51643140 | 1.50 |
ENSRNOT00000006646
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr1_-_69835185 | 1.48 |
ENSRNOT00000020757
|
LOC108349745
|
spindle assembly abnormal protein 6 homolog |
| chr10_+_20320878 | 1.45 |
ENSRNOT00000009714
|
Slit3
|
slit guidance ligand 3 |
| chr1_-_178196294 | 1.45 |
ENSRNOT00000067170
|
Btbd10
|
BTB domain containing 10 |
| chr18_-_49937522 | 1.42 |
ENSRNOT00000033254
|
Zfp608
|
zinc finger protein 608 |
| chr12_+_22535672 | 1.41 |
ENSRNOT00000073046
|
AABR07035791.1
|
|
| chr9_-_10734073 | 1.40 |
ENSRNOT00000071199
|
Kdm4b
|
lysine demethylase 4B |
| chr4_+_6330466 | 1.39 |
ENSRNOT00000089582
|
E130116L18Rik
|
RIKEN cDNA E130116L18 gene |
| chr18_+_35136132 | 1.38 |
ENSRNOT00000078731
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr2_+_30664441 | 1.36 |
ENSRNOT00000061166
|
Ak6
|
adenylate kinase 6 |
| chr4_+_109477920 | 1.33 |
ENSRNOT00000008468
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chr15_+_32355565 | 1.32 |
ENSRNOT00000072382
|
AABR07017900.1
|
|
| chr10_+_85646633 | 1.30 |
ENSRNOT00000086608
|
Psmb3
|
proteasome subunit beta 3 |
| chr1_-_91296656 | 1.27 |
ENSRNOT00000028703
|
Cebpg
|
CCAAT/enhancer binding protein gamma |
| chr9_+_88494676 | 1.24 |
ENSRNOT00000089451
|
LOC102556337
|
mitochondrial fission factor-like |
| chr1_+_168196862 | 1.23 |
ENSRNOT00000021006
|
Olr74
|
olfactory receptor 74 |
| chr13_+_90532326 | 1.23 |
ENSRNOT00000008944
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr3_+_150574473 | 1.21 |
ENSRNOT00000023905
|
Asip
|
agouti signaling protein |
| chr7_-_123259636 | 1.19 |
ENSRNOT00000007424
|
Desi1
|
desumoylating isopeptidase 1 |
| chr6_+_76174452 | 1.19 |
ENSRNOT00000084062
|
Psma6
|
proteasome subunit alpha 6 |
| chr1_+_42170281 | 1.18 |
ENSRNOT00000092879
|
Vip
|
vasoactive intestinal peptide |
| chr1_+_62964901 | 1.16 |
ENSRNOT00000075013
|
LOC100365824
|
vomeronasal 2 receptor, 22-like |
| chr16_-_81945127 | 1.15 |
ENSRNOT00000023352
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr7_+_13013541 | 1.13 |
ENSRNOT00000042792
ENSRNOT00000010549 |
Theg
|
theg spermatid protein |
| chr4_-_103403976 | 1.13 |
ENSRNOT00000088153
|
AABR07061057.3
|
|
| chr3_-_78193481 | 1.08 |
ENSRNOT00000008419
|
Olr696
|
olfactory receptor 696 |
| chr5_-_57845509 | 1.07 |
ENSRNOT00000035541
|
Kif24
|
kinesin family member 24 |
| chr10_+_49368314 | 1.05 |
ENSRNOT00000004392
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr3_+_95715193 | 1.01 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr2_-_189602143 | 1.00 |
ENSRNOT00000032062
|
Creb3l4
|
cAMP responsive element binding protein 3-like 4 |
| chr1_-_148847662 | 0.99 |
ENSRNOT00000090329
|
LOC100911376
|
olfactory receptor 226-like |
| chr2_+_30664639 | 0.99 |
ENSRNOT00000076372
ENSRNOT00000076294 ENSRNOT00000076434 ENSRNOT00000076484 |
Taf9
|
TATA-box binding protein associated factor 9 |
| chr13_+_84588702 | 0.91 |
ENSRNOT00000058689
|
Tada1
|
transcriptional adaptor 1 |
| chr3_+_15749691 | 0.87 |
ENSRNOT00000030984
|
Olr396
|
olfactory receptor 396 |
| chr10_+_43768708 | 0.87 |
ENSRNOT00000086218
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr20_+_27129291 | 0.86 |
ENSRNOT00000031053
|
AABR07044933.1
|
|
| chr1_-_78333321 | 0.86 |
ENSRNOT00000020402
|
Sae1
|
SUMO1 activating enzyme subunit 1 |
| chr1_-_226572349 | 0.85 |
ENSRNOT00000028028
|
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr5_+_164950917 | 0.85 |
ENSRNOT00000083678
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr7_-_16036185 | 0.83 |
ENSRNOT00000047899
|
LOC108348036
|
olfactory receptor 6C2-like |
| chr7_+_3900228 | 0.81 |
ENSRNOT00000071350
|
Olr921
|
olfactory receptor 921 |
| chr3_+_172195844 | 0.81 |
ENSRNOT00000034915
|
Npepl1
|
aminopeptidase-like 1 |
| chr2_+_30664217 | 0.78 |
ENSRNOT00000076786
|
Ak6
|
adenylate kinase 6 |
| chrX_-_152932953 | 0.74 |
ENSRNOT00000078929
|
Cetn2
|
centrin 2 |
| chr2_-_140334912 | 0.73 |
ENSRNOT00000015067
|
Elf2
|
E74-like factor 2 |
| chr1_-_171060011 | 0.72 |
ENSRNOT00000042829
|
Olr226
|
olfactory receptor 226 |
| chr18_+_62852303 | 0.72 |
ENSRNOT00000087673
|
Gnal
|
G protein subunit alpha L |
| chr3_-_37445907 | 0.72 |
ENSRNOT00000006172
|
Rbm43
|
RNA binding motif protein 43 |
| chr16_+_61926586 | 0.71 |
ENSRNOT00000058884
|
AABR07026059.1
|
|
| chr3_-_73580594 | 0.67 |
ENSRNOT00000012929
|
Olr488
|
olfactory receptor 488 |
| chr18_+_35249137 | 0.65 |
ENSRNOT00000017489
|
LOC100910942
|
serine protease inhibitor Kazal-type 6-like |
| chr1_-_178195948 | 0.60 |
ENSRNOT00000081767
|
Btbd10
|
BTB domain containing 10 |
| chr6_-_91456696 | 0.60 |
ENSRNOT00000005577
|
Rps29
|
ribosomal protein S29 |
| chr7_+_122160171 | 0.58 |
ENSRNOT00000074499
|
LOC100362980
|
CG3918-like |
| chr16_+_6542216 | 0.55 |
ENSRNOT00000021475
|
Dcp1a
|
decapping mRNA 1A |
| chr5_-_57168610 | 0.55 |
ENSRNOT00000090499
|
B4galt1
|
beta-1,4-galactosyltransferase 1 |
| chr7_-_5818625 | 0.53 |
ENSRNOT00000051699
|
Olr886
|
olfactory receptor 886 |
| chr14_-_2371883 | 0.51 |
ENSRNOT00000000077
|
Pde6b
|
phosphodiesterase 6B |
| chr9_+_67130259 | 0.50 |
ENSRNOT00000023796
|
Cyp20a1
|
cytochrome P450, family 20, subfamily a, polypeptide 1 |
| chr1_+_144070754 | 0.50 |
ENSRNOT00000079989
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr3_-_73401074 | 0.50 |
ENSRNOT00000012848
|
Olr476
|
olfactory receptor 476 |
| chr7_-_3707226 | 0.49 |
ENSRNOT00000064380
|
Olr879
|
olfactory receptor 879 |
| chr3_+_76890792 | 0.49 |
ENSRNOT00000087552
|
Olr639
|
olfactory receptor 639 |
| chr10_-_89084885 | 0.49 |
ENSRNOT00000027452
|
Plekhh3
|
pleckstrin homology, MyTH4 and FERM domain containing H3 |
| chr19_-_29802083 | 0.48 |
ENSRNOT00000024755
|
AC123471.1
|
|
| chr9_-_15393131 | 0.44 |
ENSRNOT00000018523
ENSRNOT00000088314 |
Med20
|
mediator complex subunit 20 |
| chr10_-_98124743 | 0.44 |
ENSRNOT00000037415
|
RGD1559578
|
RGD1559578 |
| chr3_-_21131492 | 0.42 |
ENSRNOT00000010493
|
Olr424
|
olfactory receptor 424 |
| chr6_-_26318568 | 0.40 |
ENSRNOT00000072707
|
LOC102556504
|
titin-like |
| chr1_-_142759882 | 0.40 |
ENSRNOT00000015218
|
Sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr7_+_13698647 | 0.40 |
ENSRNOT00000067342
|
Olr1086
|
olfactory receptor 1086 |
| chr16_-_19637609 | 0.40 |
ENSRNOT00000021742
|
Zfp709
|
zinc finger protein 709 |
| chr19_-_41433346 | 0.39 |
ENSRNOT00000022952
|
Cmtr2
|
cap methyltransferase 2 |
| chr8_-_113689681 | 0.34 |
ENSRNOT00000056435
|
LOC688828
|
similar to Nucleoside diphosphate-linked moiety X motif 16 (Nudix motif 16) |
| chr6_+_108796182 | 0.34 |
ENSRNOT00000006297
|
Fcf1
|
FCF1 rRNA-processing protein |
| chr20_-_2767240 | 0.33 |
ENSRNOT00000073990
|
Btnl7
|
butyrophilin-like 7 |
| chr4_-_39711271 | 0.33 |
ENSRNOT00000029602
|
Vwde
|
von Willebrand factor D and EGF domains |
| chr1_+_169410115 | 0.32 |
ENSRNOT00000023043
|
Trim6
|
tripartite motif containing 6 |
| chr10_+_34662186 | 0.32 |
ENSRNOT00000049754
|
Olr1393
|
olfactory receptor 1393 |
| chr8_-_17833773 | 0.31 |
ENSRNOT00000049758
|
Olr1117
|
olfactory receptor 1117 |
| chr14_+_45982244 | 0.30 |
ENSRNOT00000076881
ENSRNOT00000002984 |
Rell1
|
RELT-like 1 |
| chr18_+_30895831 | 0.28 |
ENSRNOT00000026985
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr10_-_92602082 | 0.28 |
ENSRNOT00000007963
|
Cdc27
|
cell division cycle 27 |
| chr1_-_78968329 | 0.26 |
ENSRNOT00000023078
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr1_-_80630038 | 0.25 |
ENSRNOT00000025281
|
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr1_-_217720414 | 0.24 |
ENSRNOT00000078810
|
Ppfia1
|
PTPRF interacting protein alpha 1 |
| chr20_+_42966140 | 0.19 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr20_-_11546046 | 0.19 |
ENSRNOT00000068374
|
LOC690386
|
hypothetical protein LOC690386 |
| chr1_-_64099277 | 0.19 |
ENSRNOT00000084846
|
Tsen34
|
tRNA splicing endonuclease subunit 34 |
| chr17_-_10559627 | 0.19 |
ENSRNOT00000023515
|
Higd2a
|
HIG1 hypoxia inducible domain family, member 2A |
| chr10_+_34553284 | 0.16 |
ENSRNOT00000049989
|
Olr1387
|
olfactory receptor 1387 |
| chr13_-_70299733 | 0.15 |
ENSRNOT00000044235
|
Smg7
|
SMG7 nonsense mediated mRNA decay factor |
| chr4_+_106361762 | 0.13 |
ENSRNOT00000052417
|
AABR07061136.1
|
|
| chr15_-_28721127 | 0.11 |
ENSRNOT00000017720
|
Mettl3
|
methyltransferase-like 3 |
| chr3_-_64024205 | 0.10 |
ENSRNOT00000037015
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr15_-_28446785 | 0.08 |
ENSRNOT00000015439
|
Olr1638
|
olfactory receptor 1638 |
| chr10_+_34564009 | 0.08 |
ENSRNOT00000041536
|
Olr1388
|
olfactory receptor 1388 |
| chr17_-_17872573 | 0.06 |
ENSRNOT00000082733
|
Rnf144b
|
ring finger protein 144B |
| chr1_-_242083484 | 0.05 |
ENSRNOT00000065921
|
Tjp2
|
tight junction protein 2 |
| chr5_+_154668179 | 0.05 |
ENSRNOT00000015971
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr17_-_89780691 | 0.05 |
ENSRNOT00000091830
|
LOC108348042
|
ankyrin repeat domain-containing protein 7-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Brca1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 2.5 | 7.5 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of establishment of T cell polarity(GO:1903903) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 1.4 | 7.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 1.2 | 4.7 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 1.0 | 5.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.8 | 3.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.8 | 2.4 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.7 | 2.9 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.6 | 3.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.6 | 4.8 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.5 | 1.5 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.5 | 1.5 | GO:0070212 | peptidyl-serine ADP-ribosylation(GO:0018312) myofibroblast differentiation(GO:0036446) protein poly-ADP-ribosylation(GO:0070212) regulation of myofibroblast differentiation(GO:1904760) regulation of interleukin-17 secretion(GO:1905076) negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.4 | 7.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.4 | 3.2 | GO:0061193 | taste bud development(GO:0061193) |
| 0.4 | 2.0 | GO:0061428 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.4 | 1.5 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.3 | 1.4 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.3 | 1.0 | GO:0021917 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.3 | 1.2 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.3 | 0.9 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.3 | 1.9 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.7 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.2 | 0.8 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 1.6 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 1.4 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.2 | 1.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 2.0 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 8.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 1.2 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.1 | 0.5 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 1.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.4 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 1.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 2.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 2.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 1.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 1.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.7 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 2.5 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 0.3 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 0.6 | GO:1903608 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 3.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 2.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 2.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 2.2 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 0.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 13.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 2.1 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 1.8 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.2 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 1.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.9 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 7.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:1904690 | primary miRNA processing(GO:0031053) regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.5 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 1.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 2.0 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 1.1 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.0 | 1.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.2 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 9.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 1.3 | 7.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.5 | 2.4 | GO:0005940 | septin ring(GO:0005940) |
| 0.3 | 1.9 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.3 | 2.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 0.9 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 0.7 | GO:0071942 | XPC complex(GO:0071942) |
| 0.2 | 1.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 7.1 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 5.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 3.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 2.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 0.8 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.2 | 1.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 1.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 1.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 1.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 5.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 3.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 9.7 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 3.5 | GO:0044445 | cytosolic part(GO:0044445) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.0 | 3.8 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.9 | 9.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.7 | 4.8 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.5 | 1.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.5 | 7.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.5 | 2.9 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 2.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.4 | 1.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 3.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 2.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 1.2 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.2 | 1.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 2.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 0.5 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.2 | 1.5 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 3.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 3.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 1.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.9 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 3.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 2.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 5.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 4.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 2.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 9.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 4.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 3.7 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 2.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 1.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 1.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 4.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.2 | 10.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 4.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.8 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 4.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.0 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 2.5 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 8.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 4.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |