Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Bhlhe40
Z-value: 0.37
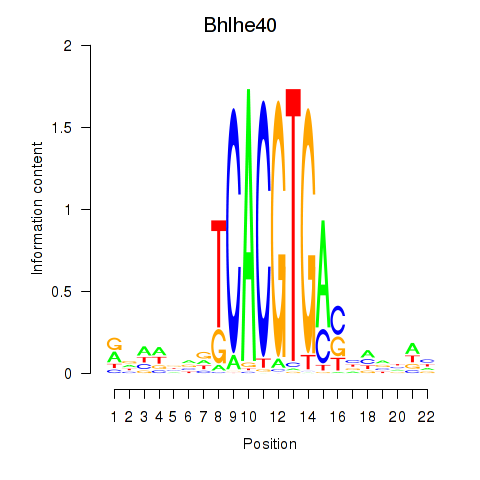
Transcription factors associated with Bhlhe40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bhlhe40
|
ENSRNOG00000007152 | basic helix-loop-helix family, member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bhlhe40 | rn6_v1_chr4_+_140703619_140703619 | -0.11 | 4.6e-02 | Click! |
Activity profile of Bhlhe40 motif
Sorted Z-values of Bhlhe40 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_106558366 | 9.36 |
ENSRNOT00000042126
|
Bex2
|
brain expressed X-linked 2 |
| chrX_-_105622156 | 8.29 |
ENSRNOT00000029511
|
Armcx2
|
armadillo repeat containing, X-linked 2 |
| chr5_+_152681101 | 7.86 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr10_+_40543288 | 7.60 |
ENSRNOT00000016755
|
Slc36a1
|
solute carrier family 36 member 1 |
| chrX_-_105417323 | 6.84 |
ENSRNOT00000015494
|
Gla
|
galactosidase, alpha |
| chr5_-_14356692 | 5.31 |
ENSRNOT00000085654
|
Atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr19_+_26151900 | 5.18 |
ENSRNOT00000005571
|
Tnpo2
|
transportin 2 |
| chr15_-_27855999 | 5.11 |
ENSRNOT00000013225
|
Tmem55b
|
transmembrane protein 55B |
| chr9_+_94425252 | 4.82 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr16_+_8497569 | 4.82 |
ENSRNOT00000027054
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chrX_+_106823491 | 4.77 |
ENSRNOT00000045997
|
Bex3
|
brain expressed X-linked 3 |
| chr5_-_164844586 | 4.61 |
ENSRNOT00000011287
|
Clcn6
|
chloride voltage-gated channel 6 |
| chrX_+_74200972 | 4.34 |
ENSRNOT00000076956
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chrX_+_74205842 | 3.92 |
ENSRNOT00000077003
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr19_-_43215077 | 3.79 |
ENSRNOT00000082151
|
Aars
|
alanyl-tRNA synthetase |
| chr2_+_93518806 | 3.38 |
ENSRNOT00000050987
|
Snx16
|
sorting nexin 16 |
| chr12_+_38459832 | 3.15 |
ENSRNOT00000090343
|
Vps33a
|
VPS33A CORVET/HOPS core subunit |
| chr6_-_80334522 | 3.00 |
ENSRNOT00000059316
|
Fbxo33
|
F-box protein 33 |
| chr4_+_99937558 | 2.91 |
ENSRNOT00000050249
|
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr10_+_94988362 | 2.82 |
ENSRNOT00000066525
|
Cep95
|
centrosomal protein 95 |
| chr19_-_43215281 | 2.78 |
ENSRNOT00000025052
|
Aars
|
alanyl-tRNA synthetase |
| chr11_+_60613882 | 2.76 |
ENSRNOT00000002853
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr13_+_84588702 | 2.76 |
ENSRNOT00000058689
|
Tada1
|
transcriptional adaptor 1 |
| chrX_-_84821775 | 2.72 |
ENSRNOT00000000174
|
Chm
|
CHM, Rab escort protein 1 |
| chr10_+_10725819 | 2.67 |
ENSRNOT00000004159
|
Glyr1
|
glyoxylate reductase 1 homolog |
| chr13_+_70174936 | 2.66 |
ENSRNOT00000064068
ENSRNOT00000079861 ENSRNOT00000092562 |
Arpc5
|
actin related protein 2/3 complex, subunit 5 |
| chr4_+_148139528 | 2.60 |
ENSRNOT00000092594
ENSRNOT00000015620 ENSRNOT00000092613 |
Washc2c
|
WASH complex subunit 2C |
| chr13_-_111765944 | 2.55 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr1_-_219450451 | 2.54 |
ENSRNOT00000025317
|
Rad9a
|
RAD9 checkpoint clamp component A |
| chr3_+_163570532 | 2.25 |
ENSRNOT00000010054
|
Arfgef2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr14_-_44767120 | 2.11 |
ENSRNOT00000003991
|
Wdr19
|
WD repeat domain 19 |
| chr1_-_101457126 | 1.75 |
ENSRNOT00000087061
ENSRNOT00000028328 |
Bax
|
BCL2 associated X, apoptosis regulator |
| chr18_+_3597240 | 1.66 |
ENSRNOT00000017045
|
RGD1311805
|
similar to RIKEN cDNA 2400010D15 |
| chr10_-_94988461 | 1.64 |
ENSRNOT00000048490
|
Ddx5
|
DEAD-box helicase 5 |
| chr1_-_164101578 | 1.56 |
ENSRNOT00000022176
|
Uvrag
|
UV radiation resistance associated |
| chr6_-_122239614 | 1.47 |
ENSRNOT00000005015
|
Galc
|
galactosylceramidase |
| chr7_+_41114697 | 1.43 |
ENSRNOT00000041354
|
Atp2b1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr6_+_109617355 | 1.35 |
ENSRNOT00000011599
|
Flvcr2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr8_-_104995725 | 1.35 |
ENSRNOT00000037120
|
Slc25a36
|
solute carrier family 25 member 36 |
| chr4_-_87257667 | 1.15 |
ENSRNOT00000083491
|
Nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr5_-_152227677 | 1.15 |
ENSRNOT00000020051
|
Dhdds
|
dehydrodolichyl diphosphate synthase subunit |
| chr2_+_198852500 | 1.05 |
ENSRNOT00000000110
|
Rnf115
|
ring finger protein 115 |
| chr2_-_196270826 | 0.99 |
ENSRNOT00000028609
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1, alpha |
| chr7_+_53878610 | 0.97 |
ENSRNOT00000091910
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr1_-_72329856 | 0.85 |
ENSRNOT00000021391
|
U2af2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr1_+_200167169 | 0.84 |
ENSRNOT00000027665
|
Sec23ip
|
SEC23 interacting protein |
| chr2_-_198852161 | 0.80 |
ENSRNOT00000028815
|
Polr3c
|
RNA polymerase III subunit C |
| chr12_+_2054680 | 0.74 |
ENSRNOT00000001290
|
Mcoln1
|
mucolipin 1 |
| chr7_+_143882000 | 0.72 |
ENSRNOT00000017110
ENSRNOT00000091053 |
Mfsd5
|
major facilitator superfamily domain containing 5 |
| chr6_+_109617596 | 0.71 |
ENSRNOT00000085031
ENSRNOT00000089972 ENSRNOT00000082402 |
Flvcr2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr5_-_160352927 | 0.70 |
ENSRNOT00000017247
|
Dnajc16
|
DnaJ heat shock protein family (Hsp40) member C16 |
| chr11_-_60613718 | 0.70 |
ENSRNOT00000002906
|
Atg3
|
autophagy related 3 |
| chr10_-_56850085 | 0.65 |
ENSRNOT00000025767
|
Rnasek
|
ribonuclease K |
| chr1_-_92119951 | 0.64 |
ENSRNOT00000018153
ENSRNOT00000092121 |
Zfp507
|
zinc finger protein 507 |
| chr2_-_210088949 | 0.60 |
ENSRNOT00000070994
|
Rbm15
|
RNA binding motif protein 15 |
| chr13_-_53870428 | 0.60 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr10_-_62287189 | 0.58 |
ENSRNOT00000004365
|
Wdr81
|
WD repeat domain 81 |
| chr1_-_282170017 | 0.57 |
ENSRNOT00000066947
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr18_+_15298978 | 0.46 |
ENSRNOT00000021263
|
Trappc8
|
trafficking protein particle complex 8 |
| chr10_-_10725655 | 0.35 |
ENSRNOT00000061236
|
Ubn1
|
ubinuclein 1 |
| chr1_-_102849430 | 0.35 |
ENSRNOT00000086856
|
Saa4
|
serum amyloid A4 |
| chr18_+_55797198 | 0.29 |
ENSRNOT00000026334
ENSRNOT00000026394 |
Dctn4
|
dynactin subunit 4 |
| chr1_-_13175876 | 0.27 |
ENSRNOT00000084870
|
Abracl
|
ABRA C-terminal like |
| chr11_+_68198709 | 0.23 |
ENSRNOT00000003048
|
Dirc2
|
disrupted in renal carcinoma 2 |
| chr1_+_144601410 | 0.13 |
ENSRNOT00000047408
|
Efl1
|
elongation factor like GTPase 1 |
| chr10_-_62191512 | 0.13 |
ENSRNOT00000090262
|
Rpa1
|
replication protein A1 |
| chr17_+_54280851 | 0.07 |
ENSRNOT00000024022
|
Arhgap12
|
Rho GTPase activating protein 12 |
| chr19_+_25803262 | 0.05 |
ENSRNOT00000003941
|
Trmt1
|
tRNA methyltransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Bhlhe40
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.9 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.6 | 6.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 1.0 | 8.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.9 | 7.6 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.7 | 5.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.7 | 2.6 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.6 | 3.1 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.6 | 1.7 | GO:0019740 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.5 | 2.1 | GO:0061055 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) myotome development(GO:0061055) |
| 0.4 | 1.6 | GO:0097680 | maintenance of Golgi location(GO:0051684) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.4 | 1.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.3 | 2.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 2.7 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.2 | 1.0 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.2 | 1.4 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.2 | 4.8 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.2 | 2.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 1.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 3.8 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.2 | 5.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 2.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 3.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 4.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.2 | 0.5 | GO:0032258 | CVT pathway(GO:0032258) |
| 0.1 | 0.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 2.9 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 1.4 | GO:1901679 | pyrimidine-containing compound transmembrane transport(GO:0072531) nucleotide transmembrane transport(GO:1901679) |
| 0.1 | 1.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 2.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 4.6 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 1.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.7 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 4.8 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 2.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.0 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 2.8 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 9.4 | GO:0051726 | regulation of cell cycle(GO:0051726) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 1.0 | 4.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.7 | 2.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.4 | 4.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 2.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.4 | 3.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 2.2 | GO:0005879 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.4 | 2.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 3.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.3 | 2.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 1.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 2.8 | GO:0030914 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.2 | 2.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 1.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.8 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 1.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 2.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 11.9 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.7 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.8 | GO:0097038 | endoplasmic reticulum exit site(GO:0070971) perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 4.8 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 2.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 7.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 4.2 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 1.6 | 6.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 1.6 | 4.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 1.2 | 4.8 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 1.0 | 5.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.9 | 6.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 2.7 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 1.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.3 | 5.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 4.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 2.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 2.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 2.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 0.7 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 2.8 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 1.6 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.2 | 5.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 4.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 2.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.9 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 1.0 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.8 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.4 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 1.1 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 1.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 2.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 2.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 6.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 1.5 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 1.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.2 | 6.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 8.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 2.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.7 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |