Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
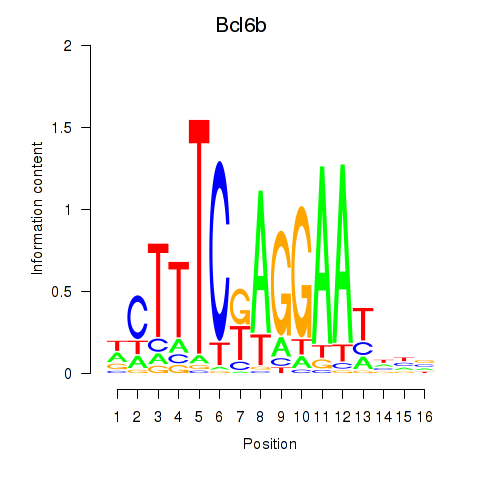
Results for Bcl6b
Z-value: 0.73
Transcription factors associated with Bcl6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bcl6b
|
ENSRNOG00000059956 | B-cell CLL/lymphoma 6B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl6b | rn6_v1_chr10_-_56839437_56839437 | -0.17 | 2.7e-03 | Click! |
Activity profile of Bcl6b motif
Sorted Z-values of Bcl6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_11585078 | 24.79 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chrX_+_20423401 | 18.14 |
ENSRNOT00000093162
|
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr11_-_11585765 | 17.07 |
ENSRNOT00000066439
|
Robo2
|
roundabout guidance receptor 2 |
| chr2_-_211322719 | 16.52 |
ENSRNOT00000027493
|
RGD1310209
|
similar to KIAA1324 protein |
| chr12_-_38782010 | 16.16 |
ENSRNOT00000001813
|
Wdr66
|
WD repeat domain 66 |
| chr4_-_17594598 | 16.00 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr2_+_208749996 | 14.77 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr5_-_79874671 | 14.63 |
ENSRNOT00000084563
|
Tnc
|
tenascin C |
| chr2_+_208750356 | 14.29 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr16_-_18766174 | 14.28 |
ENSRNOT00000084813
|
Sftpd
|
surfactant protein D |
| chr4_+_70252366 | 14.01 |
ENSRNOT00000073039
|
Chl1
|
cell adhesion molecule L1-like |
| chr14_+_17534412 | 13.11 |
ENSRNOT00000079304
ENSRNOT00000046771 |
Cdkl2
|
cyclin dependent kinase like 2 |
| chr13_+_100045343 | 12.26 |
ENSRNOT00000075792
|
LOC690288
|
similar to Dynein heavy chain at 16F CG7092-PA |
| chr16_+_39145230 | 12.17 |
ENSRNOT00000092942
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr13_+_112031594 | 12.03 |
ENSRNOT00000008440
|
Lamb3
|
laminin subunit beta 3 |
| chr2_+_54660722 | 11.88 |
ENSRNOT00000060376
|
Mroh2b
|
maestro heat-like repeat family member 2B |
| chr3_-_60611924 | 11.32 |
ENSRNOT00000068745
|
Chn1
|
chimerin 1 |
| chr16_+_6078122 | 10.91 |
ENSRNOT00000021407
|
Chdh
|
choline dehydrogenase |
| chr8_-_13513337 | 10.34 |
ENSRNOT00000071532
|
LOC108348070
|
lysine-specific demethylase 4D |
| chr16_+_39353283 | 9.63 |
ENSRNOT00000080125
|
LOC108348202
|
disintegrin and metalloproteinase domain-containing protein 21-like |
| chr13_-_36101411 | 8.52 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr8_+_28454962 | 8.12 |
ENSRNOT00000051573
|
Spata19
|
spermatogenesis associated 19 |
| chr10_+_4951557 | 7.45 |
ENSRNOT00000003451
|
Prm3
|
protamine 3 |
| chr3_-_104502471 | 7.36 |
ENSRNOT00000040306
|
Ryr3
|
ryanodine receptor 3 |
| chr8_+_115213471 | 7.18 |
ENSRNOT00000017570
|
Iqcf5
|
IQ motif containing F5 |
| chrX_+_159505344 | 6.91 |
ENSRNOT00000001164
|
Brs3
|
bombesin receptor subtype 3 |
| chr9_-_54484533 | 6.70 |
ENSRNOT00000083514
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr19_+_41482728 | 6.57 |
ENSRNOT00000022943
|
Calb2
|
calbindin 2 |
| chr1_-_13341952 | 6.53 |
ENSRNOT00000079531
|
NEWGENE_2319083
|
epithelial cell transforming 2 like |
| chr2_+_182006242 | 6.32 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr16_+_74886719 | 6.30 |
ENSRNOT00000089265
|
Atp7b
|
ATPase copper transporting beta |
| chr1_-_213921208 | 6.21 |
ENSRNOT00000044393
|
Ano9
|
anoctamin 9 |
| chr18_+_30527705 | 6.02 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr1_+_99532568 | 5.86 |
ENSRNOT00000077073
|
Zfp819
|
zinc finger protein 819 |
| chr16_+_10267482 | 5.71 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chr9_+_64898459 | 5.70 |
ENSRNOT00000029526
|
Sgo2
|
shugoshin 2 |
| chr2_+_211546560 | 5.49 |
ENSRNOT00000033443
|
Aknad1
|
AKNA domain containing 1 |
| chr7_+_27248115 | 5.47 |
ENSRNOT00000031946
ENSRNOT00000059545 |
LOC362863
|
first gene upstream of Nt5dc3 |
| chr8_+_59900651 | 4.96 |
ENSRNOT00000020410
|
Tmem266
|
transmembrane protein 266 |
| chr3_+_149261333 | 4.93 |
ENSRNOT00000038684
|
AABR07054352.1
|
|
| chr2_-_165600748 | 4.81 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chr2_-_116372226 | 4.55 |
ENSRNOT00000011932
|
Lrriq4
|
leucine-rich repeats and IQ motif containing 4 |
| chr1_+_225037737 | 4.53 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr9_+_116652530 | 4.50 |
ENSRNOT00000029210
|
L3mbtl4
|
l(3)mbt-like 4 (Drosophila) |
| chr9_+_43834310 | 4.40 |
ENSRNOT00000085720
|
Cnga3
|
cyclic nucleotide gated channel alpha 3 |
| chr16_-_6077978 | 4.02 |
ENSRNOT00000020823
|
Il17rb
|
interleukin 17 receptor B |
| chr9_-_4327679 | 3.91 |
ENSRNOT00000073468
|
LOC100910235
|
sulfotransferase 1C1-like |
| chr2_+_144646308 | 3.57 |
ENSRNOT00000078337
ENSRNOT00000093407 |
Dclk1
|
doublecortin-like kinase 1 |
| chr7_-_15364203 | 3.52 |
ENSRNOT00000036473
|
RGD1310212
|
similar to KIAA1111-like protein |
| chr8_-_38154344 | 3.45 |
ENSRNOT00000068043
|
LOC100360095
|
urinary protein 1-like |
| chr4_+_136512201 | 3.32 |
ENSRNOT00000051645
|
Cntn6
|
contactin 6 |
| chr7_-_94563001 | 3.29 |
ENSRNOT00000051139
ENSRNOT00000005561 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr14_+_80403001 | 3.24 |
ENSRNOT00000012109
|
Cpz
|
carboxypeptidase Z |
| chr12_-_24578761 | 3.17 |
ENSRNOT00000076209
|
Tbl2
|
transducin (beta)-like 2 |
| chr2_+_244058186 | 3.14 |
ENSRNOT00000021381
|
Tspan5
|
tetraspanin 5 |
| chr9_+_92565366 | 3.04 |
ENSRNOT00000023184
|
Csprs
|
component of Sp100-rs |
| chr10_-_11935382 | 3.03 |
ENSRNOT00000010059
|
Zfp174
|
zinc finger protein 174 |
| chr14_-_70098021 | 3.03 |
ENSRNOT00000004912
|
Med28
|
mediator complex subunit 28 |
| chr1_-_81412251 | 2.99 |
ENSRNOT00000026946
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr9_+_84410972 | 2.62 |
ENSRNOT00000019724
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr3_+_134440195 | 2.47 |
ENSRNOT00000072928
|
AABR07054000.1
|
|
| chr11_+_69739384 | 2.45 |
ENSRNOT00000016340
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr1_-_16687817 | 2.43 |
ENSRNOT00000091376
ENSRNOT00000081620 |
Myb
|
MYB proto-oncogene, transcription factor |
| chr9_+_81518584 | 2.33 |
ENSRNOT00000084309
|
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr10_-_13515448 | 2.25 |
ENSRNOT00000067660
|
Pdpk1
|
3-phosphoinositide dependent protein kinase-1 |
| chr8_-_38549268 | 2.25 |
ENSRNOT00000088001
|
LOC100912026
|
urinary protein 3-like |
| chr2_+_34374158 | 2.17 |
ENSRNOT00000016997
|
Adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr1_+_64506735 | 2.15 |
ENSRNOT00000086331
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
| chr10_+_106991935 | 2.07 |
ENSRNOT00000004007
|
Pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr2_+_189413966 | 2.02 |
ENSRNOT00000056603
|
RGD1564171
|
RGD1564171 |
| chr1_+_42169501 | 2.01 |
ENSRNOT00000025477
ENSRNOT00000092791 |
Vip
|
vasoactive intestinal peptide |
| chr10_-_103685844 | 1.97 |
ENSRNOT00000064284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chr6_-_94980004 | 1.97 |
ENSRNOT00000006373
|
Rtn1
|
reticulon 1 |
| chr4_+_45120576 | 1.96 |
ENSRNOT00000077961
|
ST7
|
suppression of tumorigenicity 7 |
| chrX_-_70835089 | 1.95 |
ENSRNOT00000076351
|
Tex11
|
testis expressed 11 |
| chr9_+_115916907 | 1.83 |
ENSRNOT00000023226
|
Lama1
|
laminin subunit alpha 1 |
| chr6_+_36941596 | 1.74 |
ENSRNOT00000083383
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr18_-_60002529 | 1.65 |
ENSRNOT00000081014
ENSRNOT00000024264 ENSRNOT00000059162 |
Nars
|
asparaginyl-tRNA synthetase |
| chr2_+_115739813 | 1.65 |
ENSRNOT00000085452
|
Slc7a14
|
solute carrier family 7, member 14 |
| chr10_+_66732390 | 1.59 |
ENSRNOT00000089538
|
Nf1
|
neurofibromin 1 |
| chr19_-_37528011 | 1.56 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chrX_+_156210002 | 1.51 |
ENSRNOT00000077744
|
Olr1768
|
olfactory receptor 1768 |
| chr10_+_14240219 | 1.47 |
ENSRNOT00000020233
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr20_+_10981998 | 1.25 |
ENSRNOT00000001597
|
Rrp1
|
ribosomal RNA processing 1 |
| chr18_+_30869628 | 1.25 |
ENSRNOT00000060470
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4 |
| chr2_+_165601007 | 1.19 |
ENSRNOT00000013931
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr1_+_64740487 | 1.18 |
ENSRNOT00000081213
|
LOC103691005
|
zinc finger protein 679-like |
| chr1_-_214067657 | 1.12 |
ENSRNOT00000044390
|
LOC100911881
|
chitinase domain-containing protein 1-like |
| chr10_+_72147816 | 1.09 |
ENSRNOT00000090085
|
LOC102553386
|
SUMO-conjugating enzyme UBC9-like |
| chr5_-_75910805 | 1.08 |
ENSRNOT00000019099
|
Olr854
|
olfactory receptor 854 |
| chr12_-_650376 | 1.03 |
ENSRNOT00000001468
|
N4bp2l2
|
NEDD4 binding protein 2-like 2 |
| chr10_-_59883839 | 0.99 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr8_-_85840818 | 0.98 |
ENSRNOT00000013608
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr7_-_134722215 | 0.92 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr5_-_113532878 | 0.85 |
ENSRNOT00000010173
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr1_+_171188133 | 0.81 |
ENSRNOT00000026511
|
Olr231
|
olfactory receptor 231 |
| chr9_-_60672246 | 0.74 |
ENSRNOT00000017761
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr8_-_37026142 | 0.74 |
ENSRNOT00000060446
|
LOC100361828
|
rCG22807-like |
| chrX_-_64908682 | 0.67 |
ENSRNOT00000084107
|
Zc4h2
|
zinc finger C4H2-type containing |
| chr17_-_32953641 | 0.64 |
ENSRNOT00000023332
|
Wrnip1
|
Werner helicase interacting protein 1 |
| chr7_-_4293352 | 0.56 |
ENSRNOT00000047489
|
Olr984
|
olfactory receptor 984 |
| chr12_+_25411207 | 0.51 |
ENSRNOT00000077189
ENSRNOT00000040356 |
Gtf2i
|
general transcription factor II I |
| chr2_-_29029176 | 0.51 |
ENSRNOT00000020692
|
Fcho2
|
FCH domain only 2 |
| chr2_+_200397967 | 0.49 |
ENSRNOT00000025821
|
Reg4
|
regenerating family member 4 |
| chr1_-_168456252 | 0.48 |
ENSRNOT00000021169
|
Olr92
|
olfactory receptor 92 |
| chr1_+_150797084 | 0.47 |
ENSRNOT00000018990
|
Nox4
|
NADPH oxidase 4 |
| chr15_+_37171052 | 0.43 |
ENSRNOT00000011684
|
Zmym2
|
zinc finger MYM-type containing 2 |
| chr1_-_149603272 | 0.42 |
ENSRNOT00000045582
|
Olr13
|
olfactory receptor 13 |
| chr16_-_71237118 | 0.42 |
ENSRNOT00000066974
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr7_+_42269784 | 0.40 |
ENSRNOT00000008471
ENSRNOT00000007231 |
Kitlg
|
KIT ligand |
| chr1_-_168015148 | 0.37 |
ENSRNOT00000020691
|
Olr50
|
olfactory receptor 50 |
| chr16_+_49293243 | 0.30 |
ENSRNOT00000049830
|
AC130162.1
|
|
| chr7_-_92882068 | 0.29 |
ENSRNOT00000037809
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr7_-_7311272 | 0.25 |
ENSRNOT00000040812
|
Olr1020
|
olfactory receptor 1020 |
| chr1_-_149914606 | 0.24 |
ENSRNOT00000044773
|
Olr19
|
olfactory receptor 19 |
| chr1_-_214504569 | 0.23 |
ENSRNOT00000082221
|
Chid1
|
chitinase domain containing 1 |
| chr6_-_127886540 | 0.17 |
ENSRNOT00000057298
|
LOC299277
|
similar to serine (or cysteine) peptidase inhibitor, clade A, member 3B |
| chr1_-_65863037 | 0.10 |
ENSRNOT00000075269
|
AABR07002068.1
|
|
| chr1_+_254530121 | 0.10 |
ENSRNOT00000089627
|
AC080157.1
|
|
| chr1_-_149683357 | 0.09 |
ENSRNOT00000077975
|
Olr17
|
olfactory receptor 17 |
| chr2_-_211235440 | 0.08 |
ENSRNOT00000033015
ENSRNOT00000091896 |
Sars
|
seryl-tRNA synthetase |
| chr17_+_21984325 | 0.07 |
ENSRNOT00000088725
|
AABR07027267.1
|
|
| chr6_-_139973811 | 0.01 |
ENSRNOT00000082875
|
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Bcl6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 41.9 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 6.0 | 18.1 | GO:2000688 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 5.8 | 29.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 3.7 | 14.6 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 2.9 | 14.3 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 2.3 | 6.9 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 2.1 | 6.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 2.1 | 6.2 | GO:1903796 | negative regulation of inorganic anion transmembrane transport(GO:1903796) negative regulation of chloride transport(GO:2001226) |
| 1.9 | 5.7 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 1.7 | 6.7 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 1.4 | 16.5 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 1.4 | 10.9 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 1.3 | 6.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.1 | 3.3 | GO:1903165 | response to polycyclic arene(GO:1903165) |
| 0.9 | 2.6 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.8 | 2.4 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.8 | 4.0 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.7 | 2.2 | GO:0008592 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) regulation of Toll signaling pathway(GO:0008592) negative regulation of Toll signaling pathway(GO:0045751) |
| 0.7 | 2.0 | GO:2000426 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.6 | 2.3 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.5 | 1.6 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.5 | 4.8 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.5 | 2.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.5 | 3.9 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.5 | 17.6 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.4 | 1.9 | GO:0006311 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.4 | 4.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.3 | 1.0 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.3 | 5.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.3 | 4.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 14.0 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.3 | 11.9 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.3 | 11.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 1.8 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.3 | 1.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 2.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.2 | 8.6 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 12.0 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.2 | 3.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 6.7 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.2 | 2.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.2 | 2.3 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 6.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.7 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.9 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.1 | 11.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 3.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.6 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.4 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 3.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.0 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 7.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.2 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 1.0 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.2 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 4.6 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.0 | 1.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.7 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 6.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 5.7 | GO:0016569 | covalent chromatin modification(GO:0016569) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 12.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.2 | 41.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.9 | 1.8 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.8 | 7.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.8 | 6.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 14.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 11.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.4 | 5.7 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.4 | 4.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.4 | 2.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.4 | 1.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 1.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.3 | 14.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 4.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 6.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 6.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 0.5 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.1 | 1.9 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 18.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 1.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 16.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 1.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 7.4 | GO:0000786 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.1 | 3.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 3.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 2.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 8.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 44.5 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 3.6 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 13.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 13.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 4.6 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 1.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 29.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 3.8 | 11.3 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 3.5 | 41.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 2.3 | 16.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 2.3 | 6.9 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 1.8 | 7.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.5 | 18.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.3 | 16.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.3 | 6.3 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 1.1 | 3.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.6 | 14.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.6 | 3.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.6 | 6.7 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.5 | 4.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.5 | 2.0 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.5 | 2.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.5 | 13.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.4 | 2.6 | GO:0004144 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.4 | 2.3 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.4 | 4.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.4 | 4.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 3.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.3 | 1.0 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.3 | 6.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 2.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.2 | 2.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.2 | 1.8 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 1.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 24.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 0.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 3.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 10.3 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.1 | 3.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 10.9 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 2.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 5.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 14.0 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 3.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 6.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 7.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.7 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 2.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.6 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 9.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 3.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 12.0 | GO:0032403 | protein complex binding(GO:0032403) |
| 0.0 | 1.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 13.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 6.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.5 | 14.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 13.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 14.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 6.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 5.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 4.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 16.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 3.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 16.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.8 | 41.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.5 | 6.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 6.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 18.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 6.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 9.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 12.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 6.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |