Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
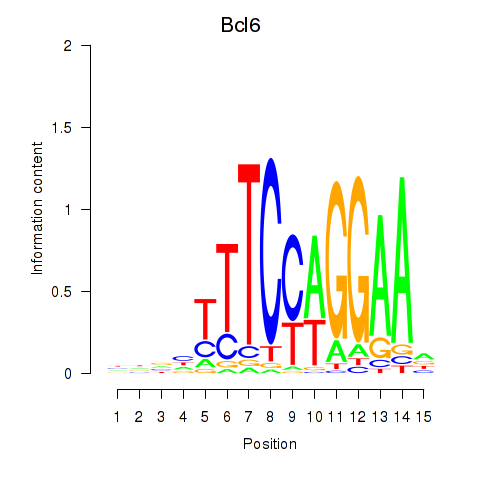
Results for Bcl6
Z-value: 1.01
Transcription factors associated with Bcl6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bcl6
|
ENSRNOG00000001843 | B-cell CLL/lymphoma 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl6 | rn6_v1_chr11_+_80255790_80255790 | -0.52 | 2.8e-23 | Click! |
Activity profile of Bcl6 motif
Sorted Z-values of Bcl6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_29538380 | 61.09 |
ENSRNOT00000010845
|
Calb1
|
calbindin 1 |
| chr1_-_89360733 | 35.41 |
ENSRNOT00000028544
|
Mag
|
myelin-associated glycoprotein |
| chr4_+_147037179 | 33.71 |
ENSRNOT00000011292
|
Syn2
|
synapsin II |
| chr17_-_10818835 | 29.83 |
ENSRNOT00000091046
|
Cplx2
|
complexin 2 |
| chr17_+_47721977 | 29.51 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr2_-_89310946 | 29.15 |
ENSRNOT00000015195
|
Ralyl
|
RALY RNA binding protein-like |
| chr8_-_80631873 | 24.64 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr2_+_60920257 | 24.32 |
ENSRNOT00000025170
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr14_+_8182383 | 23.67 |
ENSRNOT00000092719
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr18_-_37096132 | 23.13 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_+_106998623 | 22.21 |
ENSRNOT00000022383
|
Slc17a6
|
solute carrier family 17 member 6 |
| chr12_-_10307265 | 21.75 |
ENSRNOT00000092627
|
Wasf3
|
WAS protein family, member 3 |
| chr15_-_52317219 | 21.56 |
ENSRNOT00000016555
|
Dmtn
|
dematin actin binding protein |
| chr13_+_84474319 | 21.39 |
ENSRNOT00000031367
ENSRNOT00000072244 ENSRNOT00000072897 ENSRNOT00000064168 ENSRNOT00000074954 ENSRNOT00000073696 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr2_+_200793571 | 20.63 |
ENSRNOT00000091444
|
Hao2
|
hydroxyacid oxidase 2 |
| chr11_+_66932614 | 20.46 |
ENSRNOT00000003189
|
Slc15a2
|
solute carrier family 15 member 2 |
| chr14_+_17534412 | 20.43 |
ENSRNOT00000079304
ENSRNOT00000046771 |
Cdkl2
|
cyclin dependent kinase like 2 |
| chr1_+_156552328 | 20.33 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr9_-_85243001 | 20.29 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr15_+_52767442 | 19.68 |
ENSRNOT00000014441
|
LOC108348161
|
phytanoyl-CoA hydroxylase-interacting protein |
| chr4_-_55398941 | 18.48 |
ENSRNOT00000075324
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr10_-_61361250 | 18.47 |
ENSRNOT00000092203
|
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr15_+_52148379 | 17.73 |
ENSRNOT00000074912
|
Phyhip
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr3_+_56862691 | 17.72 |
ENSRNOT00000087712
|
Gad1
|
glutamate decarboxylase 1 |
| chr9_-_27452902 | 17.33 |
ENSRNOT00000018325
|
Gsta1
|
glutathione S-transferase alpha 1 |
| chr1_+_32221636 | 17.18 |
ENSRNOT00000022346
ENSRNOT00000089941 |
Slc6a18
|
solute carrier family 6 member 18 |
| chr3_+_97723901 | 17.12 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr20_+_20378861 | 16.55 |
ENSRNOT00000091044
|
Ank3
|
ankyrin 3 |
| chr7_-_76294663 | 16.39 |
ENSRNOT00000064513
|
Ncald
|
neurocalcin delta |
| chr4_-_11610518 | 16.35 |
ENSRNOT00000066643
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_+_9981958 | 16.08 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr2_-_185303610 | 15.99 |
ENSRNOT00000093479
ENSRNOT00000046286 |
NEWGENE_1592020
|
protease, serine 48 |
| chr8_+_85503224 | 15.88 |
ENSRNOT00000012348
|
Gsta4
|
glutathione S-transferase alpha 4 |
| chr2_+_58448917 | 15.10 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chrX_+_28072826 | 15.01 |
ENSRNOT00000039796
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr5_+_22392732 | 14.99 |
ENSRNOT00000087182
|
Clvs1
|
clavesin 1 |
| chrX_-_123092217 | 14.88 |
ENSRNOT00000039710
|
Gm14569
|
predicted gene 14569 |
| chr16_+_54765325 | 14.87 |
ENSRNOT00000065327
ENSRNOT00000086899 |
Mtmr7
|
myotubularin related protein 7 |
| chr5_-_2982603 | 14.40 |
ENSRNOT00000045460
|
Kcnb2
|
potassium voltage-gated channel subfamily B member 2 |
| chrX_+_20423401 | 14.40 |
ENSRNOT00000093162
|
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr17_+_53231343 | 14.21 |
ENSRNOT00000021703
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr7_+_23403891 | 13.61 |
ENSRNOT00000037918
|
Syn3
|
synapsin III |
| chr16_+_74886719 | 13.47 |
ENSRNOT00000089265
|
Atp7b
|
ATPase copper transporting beta |
| chrX_+_71199491 | 13.44 |
ENSRNOT00000076168
ENSRNOT00000005077 ENSRNOT00000005102 |
Nlgn3
|
neuroligin 3 |
| chr8_-_53899669 | 13.27 |
ENSRNOT00000082257
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr2_+_115739813 | 13.01 |
ENSRNOT00000085452
|
Slc7a14
|
solute carrier family 7, member 14 |
| chr6_+_95323579 | 12.98 |
ENSRNOT00000007369
|
Pcnx4
|
pecanex homolog 4 (Drosophila) |
| chr4_-_117568348 | 12.84 |
ENSRNOT00000071447
|
Nat8f2
|
N-acetyltransferase 8 (GCN5-related) family member 2 |
| chr16_+_23553647 | 12.81 |
ENSRNOT00000041994
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_-_75910805 | 12.65 |
ENSRNOT00000019099
|
Olr854
|
olfactory receptor 854 |
| chr3_+_177310753 | 12.52 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr8_+_127735258 | 12.32 |
ENSRNOT00000015888
|
Vill
|
villin-like |
| chr11_-_11585078 | 12.11 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr12_-_46493203 | 12.09 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr13_+_90723092 | 12.00 |
ENSRNOT00000010146
|
Kcnj10
|
ATP-sensitive inward rectifier potassium channel 10 |
| chr3_+_146546387 | 11.43 |
ENSRNOT00000009946
|
Entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 |
| chr16_-_81945127 | 10.98 |
ENSRNOT00000023352
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr7_-_134722215 | 10.98 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr10_-_59883839 | 10.83 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr16_+_74076812 | 10.69 |
ENSRNOT00000025675
|
Ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr3_-_2803574 | 10.00 |
ENSRNOT00000040995
|
RGD1560470
|
similar to Gene model 996 |
| chr16_+_75145401 | 9.99 |
ENSRNOT00000046821
ENSRNOT00000047761 |
Spag11bl
|
sperm associated antigen 11b-like |
| chr5_+_133896141 | 9.87 |
ENSRNOT00000011434
|
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr10_-_15204143 | 9.70 |
ENSRNOT00000026971
|
Rhbdl1
|
rhomboid like 1 |
| chr19_-_57333433 | 9.61 |
ENSRNOT00000024917
|
Agt
|
angiotensinogen |
| chr9_-_65790347 | 9.49 |
ENSRNOT00000028506
|
AABR07067812.1
|
|
| chr9_+_84410972 | 9.48 |
ENSRNOT00000019724
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr4_-_117575154 | 9.46 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr16_+_19097391 | 9.46 |
ENSRNOT00000018018
|
Calr3
|
calreticulin 3 |
| chr2_+_144646308 | 9.31 |
ENSRNOT00000078337
ENSRNOT00000093407 |
Dclk1
|
doublecortin-like kinase 1 |
| chr1_-_250774105 | 9.29 |
ENSRNOT00000079227
|
Sgms1
|
sphingomyelin synthase 1 |
| chr13_-_88061108 | 9.23 |
ENSRNOT00000003774
|
Rgs4
|
regulator of G-protein signaling 4 |
| chr6_+_109110534 | 9.21 |
ENSRNOT00000009156
|
Zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr7_+_6644643 | 9.12 |
ENSRNOT00000051670
|
Olr962
|
olfactory receptor 962 |
| chr1_+_225037737 | 8.87 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr1_+_81456984 | 8.70 |
ENSRNOT00000027075
|
Ethe1
|
ETHE1, persulfide dioxygenase |
| chr2_+_78245459 | 8.61 |
ENSRNOT00000089551
|
LOC103689968
|
protein FAM134B |
| chr7_-_3342491 | 8.37 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chrX_+_13441558 | 8.24 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr14_-_37294472 | 8.23 |
ENSRNOT00000081087
|
Dcun1d4
|
defective in cullin neddylation 1 domain containing 4 |
| chr7_-_8060373 | 8.20 |
ENSRNOT00000071699
|
Olfr798
|
olfactory receptor 798 |
| chr8_-_12902262 | 8.15 |
ENSRNOT00000033969
|
Endod1
|
endonuclease domain containing 1 |
| chr6_-_38228379 | 8.13 |
ENSRNOT00000084924
|
Mycn
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr8_+_21890319 | 7.95 |
ENSRNOT00000027949
|
LOC500956
|
hypothetical protein LOC500956 |
| chr10_+_90342051 | 7.75 |
ENSRNOT00000028487
|
Rundc3a
|
RUN domain containing 3A |
| chr2_+_225827504 | 7.73 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr17_+_43050961 | 7.61 |
ENSRNOT00000059510
|
Carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr18_+_30017918 | 7.48 |
ENSRNOT00000079794
|
Pcdha4
|
protocadherin alpha 4 |
| chr3_-_117990289 | 7.33 |
ENSRNOT00000011084
|
Shc4
|
SHC adaptor protein 4 |
| chr1_+_42169501 | 7.16 |
ENSRNOT00000025477
ENSRNOT00000092791 |
Vip
|
vasoactive intestinal peptide |
| chr4_+_154309426 | 7.14 |
ENSRNOT00000019346
|
A2m
|
alpha-2-macroglobulin |
| chr4_+_78080310 | 7.11 |
ENSRNOT00000035906
|
Sspo
|
SCO-spondin |
| chr5_+_139597731 | 7.00 |
ENSRNOT00000072427
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr7_+_132857628 | 6.95 |
ENSRNOT00000005438
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr4_+_154423209 | 6.88 |
ENSRNOT00000075799
|
LOC100911545
|
alpha-2-macroglobulin-like |
| chr1_-_188097530 | 6.84 |
ENSRNOT00000078825
|
Syt17
|
synaptotagmin 17 |
| chrX_+_9436707 | 6.79 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr16_+_2379480 | 6.76 |
ENSRNOT00000079215
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr14_-_92495894 | 6.58 |
ENSRNOT00000064483
|
Cobl
|
cordon-bleu WH2 repeat protein |
| chr5_+_162808646 | 6.56 |
ENSRNOT00000021155
|
Dhrs3
|
dehydrogenase/reductase 3 |
| chr14_+_115275894 | 6.52 |
ENSRNOT00000033437
|
Gpr75
|
G protein-coupled receptor 75 |
| chr17_+_15194262 | 6.40 |
ENSRNOT00000073070
|
AABR07072539.1
|
|
| chr17_+_14469488 | 6.39 |
ENSRNOT00000060670
|
AABR07027111.1
|
|
| chr16_+_54291251 | 6.26 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr9_-_61134963 | 6.14 |
ENSRNOT00000017967
|
Pgap1
|
post-GPI attachment to proteins 1 |
| chrX_+_159505344 | 6.10 |
ENSRNOT00000001164
|
Brs3
|
bombesin receptor subtype 3 |
| chr10_-_91699424 | 6.09 |
ENSRNOT00000004650
|
Lyzl6
|
lysozyme-like 6 |
| chr5_+_58855773 | 6.07 |
ENSRNOT00000072869
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr7_-_94563001 | 6.01 |
ENSRNOT00000051139
ENSRNOT00000005561 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr16_+_4997461 | 5.99 |
ENSRNOT00000071042
|
LOC102547963
|
leucine-rich repeat and transmembrane domain-containing protein 1-like |
| chr16_-_19996716 | 5.87 |
ENSRNOT00000024434
ENSRNOT00000080706 |
Nxnl1
|
nucleoredoxin-like 1 |
| chr15_+_33822730 | 5.86 |
ENSRNOT00000067607
|
RGD1564324
|
similar to dehydrogenase/reductase member 2 |
| chr8_-_63150753 | 5.73 |
ENSRNOT00000078666
|
Tbc1d21
|
TBC1 domain family, member 21 |
| chr4_-_148711830 | 5.55 |
ENSRNOT00000049091
|
Olr832
|
olfactory receptor 832 |
| chr10_+_58784660 | 5.40 |
ENSRNOT00000019925
|
LOC100912483
|
uncharacterized LOC100912483 |
| chr2_-_200003443 | 5.40 |
ENSRNOT00000024900
ENSRNOT00000088041 |
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr5_+_164845925 | 5.37 |
ENSRNOT00000011384
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr14_-_42560174 | 5.34 |
ENSRNOT00000003128
|
Tmem33
|
transmembrane protein 33 |
| chr7_-_102298522 | 5.34 |
ENSRNOT00000006273
|
A1bg
|
alpha-1-B glycoprotein |
| chr11_-_60251415 | 5.32 |
ENSRNOT00000080148
ENSRNOT00000086602 |
Slc9c1
|
solute carrier family 9 member C1 |
| chr1_-_188097374 | 5.19 |
ENSRNOT00000092246
|
Syt17
|
synaptotagmin 17 |
| chr15_+_89407426 | 5.06 |
ENSRNOT00000039423
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr9_+_51302151 | 5.00 |
ENSRNOT00000085908
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr15_-_105402708 | 4.99 |
ENSRNOT00000057564
|
Oxgr1
|
oxoglutarate receptor 1 |
| chrX_-_106066227 | 4.93 |
ENSRNOT00000033752
|
Nxf7
|
nuclear RNA export factor 7 |
| chr7_-_15163866 | 4.93 |
ENSRNOT00000042638
|
Zfp870
|
zinc finger protein 870 |
| chr11_-_54344615 | 4.92 |
ENSRNOT00000090307
|
Myh15
|
myosin, heavy chain 15 |
| chr7_-_139223116 | 4.92 |
ENSRNOT00000086559
|
Endou
|
endonuclease, poly(U)-specific |
| chr16_-_31972507 | 4.89 |
ENSRNOT00000032199
|
Cbr4
|
carbonyl reductase 4 |
| chr4_-_182600258 | 4.86 |
ENSRNOT00000067970
|
Ergic2
|
ERGIC and golgi 2 |
| chr5_+_169452493 | 4.86 |
ENSRNOT00000014444
|
Gpr153
|
G protein-coupled receptor 153 |
| chr1_-_149683357 | 4.83 |
ENSRNOT00000077975
|
Olr17
|
olfactory receptor 17 |
| chr4_-_150829913 | 4.54 |
ENSRNOT00000041571
|
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr8_-_126390801 | 4.54 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr1_-_149914606 | 4.51 |
ENSRNOT00000044773
|
Olr19
|
olfactory receptor 19 |
| chr4_-_58216711 | 4.50 |
ENSRNOT00000015134
|
Tsga13
|
testis specific, 13 |
| chr13_+_83721300 | 4.44 |
ENSRNOT00000082677
|
Adcy10
|
adenylate cyclase 10 (soluble) |
| chr10_+_92245442 | 4.42 |
ENSRNOT00000006808
|
Sppl2c
|
signal peptide peptidase like 2C |
| chr14_+_36216002 | 4.40 |
ENSRNOT00000038506
|
Scfd2
|
sec1 family domain containing 2 |
| chr9_-_63291350 | 4.35 |
ENSRNOT00000058831
|
Hsfy2
|
heat shock transcription factor, Y linked 2 |
| chrX_+_45637415 | 4.31 |
ENSRNOT00000050544
|
RGD1563378
|
similar to ferritin heavy polypeptide-like 17 |
| chr3_+_117335212 | 4.30 |
ENSRNOT00000043611
|
Slc24a5
|
solute carrier family 24 member 5 |
| chr2_-_165600748 | 4.26 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chr6_-_105160470 | 4.24 |
ENSRNOT00000008367
|
Adam4
|
a disintegrin and metalloprotease domain 4 |
| chr1_-_171028743 | 4.21 |
ENSRNOT00000026488
|
Olr223
|
olfactory receptor 223 |
| chr5_+_28395296 | 4.20 |
ENSRNOT00000009375
|
Tmem55a
|
transmembrane protein 55A |
| chr4_-_150829741 | 4.17 |
ENSRNOT00000051846
ENSRNOT00000052017 |
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr20_+_2194709 | 4.13 |
ENSRNOT00000001017
|
Trim15
|
tripartite motif containing 15 |
| chr19_+_15294248 | 4.10 |
ENSRNOT00000024622
|
Ces1f
|
carboxylesterase 1F |
| chr19_+_37282018 | 3.98 |
ENSRNOT00000021723
|
Tmem208
|
transmembrane protein 208 |
| chr7_+_5655645 | 3.92 |
ENSRNOT00000043749
|
LOC108348038
|
olfactory receptor 6C2-like |
| chr1_+_171188133 | 3.91 |
ENSRNOT00000026511
|
Olr231
|
olfactory receptor 231 |
| chr1_+_171274305 | 3.83 |
ENSRNOT00000051038
|
Olr234
|
olfactory receptor 234 |
| chr6_+_72891725 | 3.82 |
ENSRNOT00000038074
|
Nubpl
|
nucleotide binding protein-like |
| chr2_+_54660722 | 3.64 |
ENSRNOT00000060376
|
Mroh2b
|
maestro heat-like repeat family member 2B |
| chr3_-_4405194 | 3.64 |
ENSRNOT00000072025
|
AABR07051266.1
|
|
| chr12_+_16030788 | 3.62 |
ENSRNOT00000051565
|
Iqce
|
IQ motif containing E |
| chr2_+_182006242 | 3.60 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr1_+_190666149 | 3.59 |
ENSRNOT00000089361
|
LOC102547219
|
uncharacterized LOC102547219 |
| chr4_+_146455332 | 3.57 |
ENSRNOT00000009775
|
Hrh1
|
histamine receptor H 1 |
| chr16_+_54358471 | 3.48 |
ENSRNOT00000047314
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr3_+_77414999 | 3.35 |
ENSRNOT00000047910
|
Olr659
|
olfactory receptor 659 |
| chr4_-_168656673 | 3.35 |
ENSRNOT00000009341
|
Gpr19
|
G protein-coupled receptor 19 |
| chr1_-_171045419 | 3.29 |
ENSRNOT00000047239
|
Olr224
|
olfactory receptor 224 |
| chr19_-_37528011 | 3.28 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr7_-_144109116 | 3.21 |
ENSRNOT00000020437
|
Map3k12
|
mitogen activated protein kinase kinase kinase 12 |
| chr10_-_89088993 | 3.17 |
ENSRNOT00000027458
|
Ccr10
|
C-C motif chemokine receptor 10 |
| chrX_+_136488691 | 3.13 |
ENSRNOT00000093432
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr4_-_58195143 | 3.10 |
ENSRNOT00000033706
|
Copg2
|
coatomer protein complex, subunit gamma 2 |
| chr15_+_23792931 | 3.08 |
ENSRNOT00000092091
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr20_+_1736377 | 3.05 |
ENSRNOT00000047035
|
Olr1734
|
olfactory receptor 1734 |
| chr2_+_53109684 | 3.04 |
ENSRNOT00000086590
|
Selenop
|
selenoprotein P |
| chr1_-_89369960 | 3.04 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr14_+_17195014 | 3.01 |
ENSRNOT00000031667
|
Cxcl11
|
C-X-C motif chemokine ligand 11 |
| chrX_+_156854594 | 2.94 |
ENSRNOT00000083442
|
Renbp
|
renin binding protein |
| chr3_+_149261333 | 2.89 |
ENSRNOT00000038684
|
AABR07054352.1
|
|
| chr9_+_100104000 | 2.87 |
ENSRNOT00000074160
|
Capn10
|
calpain 10 |
| chr1_+_70480941 | 2.84 |
ENSRNOT00000072404
|
Olr6
|
olfactory receptor 6 |
| chr17_+_9830332 | 2.82 |
ENSRNOT00000021946
|
Rab24
|
RAB24, member RAS oncogene family |
| chr1_-_114371897 | 2.82 |
ENSRNOT00000018028
|
Nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 |
| chr7_+_139685573 | 2.77 |
ENSRNOT00000088376
|
Pfkm
|
phosphofructokinase, muscle |
| chr6_-_76652757 | 2.69 |
ENSRNOT00000042177
|
LOC100359668
|
ferritin light chain 2 |
| chr7_-_7617499 | 2.66 |
ENSRNOT00000051053
|
Olr950
|
olfactory receptor 950 |
| chr19_+_37668693 | 2.65 |
ENSRNOT00000084970
|
Pard6a
|
par-6 family cell polarity regulator alpha |
| chrX_-_71127237 | 2.65 |
ENSRNOT00000076403
ENSRNOT00000076635 ENSRNOT00000068098 |
Snx12
|
sorting nexin 12 |
| chr3_+_75379451 | 2.63 |
ENSRNOT00000071725
|
Olr558
|
olfactory receptor 558 |
| chr10_+_106991935 | 2.58 |
ENSRNOT00000004007
|
Pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr1_-_149603272 | 2.56 |
ENSRNOT00000045582
|
Olr13
|
olfactory receptor 13 |
| chr8_+_22750336 | 2.55 |
ENSRNOT00000013496
|
Ldlr
|
low density lipoprotein receptor |
| chr19_-_37281933 | 2.52 |
ENSRNOT00000059663
|
Lrrc29
|
leucine rich repeat containing 29 |
| chr2_-_259150479 | 2.46 |
ENSRNOT00000085892
|
AABR07013843.1
|
|
| chr4_+_9821541 | 2.45 |
ENSRNOT00000015733
|
Slc26a5
|
solute carrier family 26 member 5 |
| chr1_+_85337805 | 2.44 |
ENSRNOT00000026839
|
Samd4b
|
sterile alpha motif domain containing 4B |
| chr7_-_6971557 | 2.44 |
ENSRNOT00000050153
|
Olr960
|
olfactory receptor 960 |
| chr5_-_164585634 | 2.43 |
ENSRNOT00000065985
|
Mfn2
|
mitofusin 2 |
| chr7_-_70328148 | 2.41 |
ENSRNOT00000074645
|
Mettl21b
|
methyltransferase like 21B |
| chr19_+_43220508 | 2.34 |
ENSRNOT00000091788
ENSRNOT00000084462 ENSRNOT00000087474 |
Ddx19a
|
DEAD-box helicase 19A |
| chr1_-_200696928 | 2.33 |
ENSRNOT00000068183
ENSRNOT00000022253 ENSRNOT00000065448 ENSRNOT00000022331 |
Fgfr2
|
fibroblast growth factor receptor 2 |
| chr15_+_103344476 | 2.32 |
ENSRNOT00000013329
ENSRNOT00000077047 ENSRNOT00000078103 |
Gpr180
|
G protein-coupled receptor 180 |
| chr2_-_209582627 | 2.29 |
ENSRNOT00000024351
|
Olr392
|
olfactory receptor 392 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Bcl6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.3 | 61.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 6.9 | 20.6 | GO:0018924 | mandelate metabolic process(GO:0018924) |
| 5.4 | 21.6 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 5.1 | 20.5 | GO:0042891 | antibiotic transport(GO:0042891) dipeptide transport(GO:0042938) |
| 4.8 | 14.4 | GO:2000682 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 4.5 | 13.5 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 4.4 | 13.3 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 4.1 | 16.5 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 4.0 | 16.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 4.0 | 12.0 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 3.5 | 17.7 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 3.5 | 7.0 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 3.5 | 24.3 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 3.2 | 9.6 | GO:0003331 | ovarian follicle rupture(GO:0001543) angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) angiotensin-mediated drinking behavior(GO:0003051) regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) positive regulation of gap junction assembly(GO:1903598) |
| 3.2 | 9.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 3.0 | 57.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 3.0 | 29.8 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 2.5 | 7.6 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 2.5 | 22.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 2.4 | 12.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 2.3 | 29.8 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 2.2 | 24.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 2.2 | 8.7 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 2.2 | 23.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 2.0 | 6.1 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 2.0 | 6.0 | GO:1903165 | response to polycyclic arene(GO:1903165) |
| 1.9 | 7.7 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 1.9 | 15.0 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 1.8 | 20.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 1.8 | 7.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 1.7 | 8.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 1.7 | 18.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.5 | 23.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 1.5 | 8.7 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 1.4 | 4.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 1.3 | 5.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 1.3 | 17.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 1.3 | 9.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 1.3 | 6.6 | GO:0001757 | somite specification(GO:0001757) |
| 1.3 | 11.7 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 1.2 | 3.6 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 1.2 | 15.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 1.2 | 2.3 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 1.1 | 12.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 1.1 | 15.9 | GO:0009635 | response to herbicide(GO:0009635) |
| 1.0 | 8.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 1.0 | 14.9 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.9 | 7.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.9 | 5.4 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.9 | 6.1 | GO:0015791 | polyol transport(GO:0015791) |
| 0.9 | 4.3 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.8 | 2.5 | GO:0009751 | response to salicylic acid(GO:0009751) |
| 0.8 | 2.4 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.8 | 14.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.8 | 10.8 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.8 | 3.0 | GO:0034760 | negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.8 | 1.5 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.7 | 2.9 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.7 | 6.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.7 | 3.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.7 | 7.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.7 | 4.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.7 | 2.8 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.7 | 2.6 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.6 | 3.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.6 | 21.2 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.6 | 2.6 | GO:0090118 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.6 | 8.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.6 | 4.9 | GO:0030638 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.6 | 4.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.6 | 2.4 | GO:1990910 | response to hypobaric hypoxia(GO:1990910) |
| 0.5 | 7.0 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.5 | 10.7 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.5 | 12.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.5 | 2.7 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.5 | 12.0 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.5 | 1.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.5 | 33.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.5 | 1.9 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.5 | 8.8 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.5 | 1.4 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.4 | 4.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.4 | 3.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.4 | 5.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 17.6 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.4 | 11.4 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.4 | 10.0 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.4 | 14.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.4 | 2.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.4 | 8.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.4 | 4.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.3 | 8.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 25.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.3 | 15.0 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.3 | 2.6 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.2 | 3.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.2 | 4.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 1.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.2 | 9.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 2.8 | GO:0015693 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 0.2 | 3.0 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.2 | 10.2 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.2 | 3.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 4.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 4.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.2 | 1.8 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 8.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.2 | 13.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.2 | 13.6 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 2.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.8 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 4.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 3.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.6 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 3.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 16.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 5.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 2.0 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 3.6 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 12.0 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 6.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 8.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 1.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 4.9 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 0.4 | GO:0007060 | meiotic gene conversion(GO:0006311) male meiosis chromosome segregation(GO:0007060) gene conversion(GO:0035822) |
| 0.1 | 5.7 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.2 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.7 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 5.1 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 2.9 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 10.6 | GO:0007369 | gastrulation(GO:0007369) |
| 0.0 | 3.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 1.1 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 0.7 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 2.4 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.5 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.8 | 35.4 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 7.2 | 21.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 5.0 | 29.8 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 3.1 | 24.6 | GO:0044305 | calyx of Held(GO:0044305) |
| 2.1 | 16.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 1.7 | 7.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.6 | 16.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 1.3 | 20.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 1.3 | 20.3 | GO:0031045 | dense core granule(GO:0031045) |
| 1.1 | 22.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.0 | 9.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.9 | 6.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.9 | 2.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.9 | 6.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.9 | 31.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.9 | 23.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.8 | 33.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.8 | 7.6 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.7 | 8.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.7 | 12.0 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.7 | 17.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.6 | 4.4 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.6 | 6.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.6 | 13.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.5 | 1.6 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.5 | 61.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.4 | 3.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 14.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.4 | 12.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.4 | 10.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.4 | 18.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 1.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.3 | 24.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 2.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 1.6 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.3 | 3.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 13.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 3.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 15.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 15.0 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.2 | 2.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 1.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.2 | 4.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 17.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 1.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 3.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 8.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 3.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 12.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 12.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.2 | 2.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 13.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 16.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 20.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 6.8 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 28.9 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 12.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 20.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 12.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 2.4 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 9.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 3.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 7.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 25.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 5.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 2.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 4.2 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 20.5 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.5 | GO:0098800 | inner mitochondrial membrane protein complex(GO:0098800) |
| 0.0 | 1.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 4.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.5 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.3 | 61.1 | GO:0099510 | calcium ion binding involved in regulation of cytosolic calcium ion concentration(GO:0099510) |
| 6.9 | 20.6 | GO:0052854 | very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 6.8 | 20.5 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 4.6 | 18.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 4.1 | 16.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 3.6 | 10.8 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 3.5 | 17.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 3.0 | 12.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 2.9 | 11.4 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 2.7 | 24.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 2.7 | 13.5 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 2.7 | 16.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 2.5 | 35.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 2.4 | 23.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.3 | 7.0 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 2.0 | 20.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 2.0 | 6.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 2.0 | 6.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 1.9 | 9.5 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 1.9 | 9.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.8 | 33.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 1.8 | 7.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 1.6 | 4.9 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 1.5 | 8.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 1.4 | 20.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.4 | 22.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 1.2 | 14.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.1 | 7.7 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 1.1 | 29.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.0 | 12.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.0 | 9.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.9 | 40.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.9 | 2.6 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.8 | 4.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.7 | 20.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.7 | 3.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.7 | 20.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.7 | 2.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.7 | 14.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.7 | 17.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.6 | 14.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.6 | 15.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.6 | 1.9 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.6 | 14.9 | GO:0052866 | phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.6 | 4.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.6 | 16.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.6 | 3.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 33.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.5 | 5.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 12.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.5 | 2.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.5 | 10.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.5 | 8.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.5 | 13.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.5 | 6.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.4 | 7.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.4 | 3.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 23.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.3 | 1.0 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.3 | 12.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 9.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.3 | 4.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.3 | 2.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 15.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.3 | 5.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.3 | 5.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 2.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 9.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 22.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.2 | 5.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 2.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 15.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.2 | 1.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 7.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 13.0 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.2 | 4.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 0.5 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 14.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 5.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 3.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 3.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 4.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 22.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 4.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 10.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 6.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 6.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 5.5 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 1.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 6.4 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 13.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 4.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 23.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.6 | 30.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.5 | 4.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.5 | 10.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 9.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 8.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.3 | 11.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.3 | 20.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 9.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 11.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 1.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 8.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 18.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 3.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 2.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 3.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 47.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 1.5 | 16.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 1.5 | 17.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 1.4 | 40.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 1.2 | 35.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 1.2 | 59.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 1.2 | 18.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 1.1 | 18.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 1.0 | 16.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.8 | 18.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.6 | 16.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.6 | 16.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.4 | 12.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.4 | 7.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 13.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.3 | 7.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 13.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 14.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 12.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 3.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 7.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 2.6 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.2 | 6.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 2.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.2 | 5.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 19.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 1.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 4.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |