Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
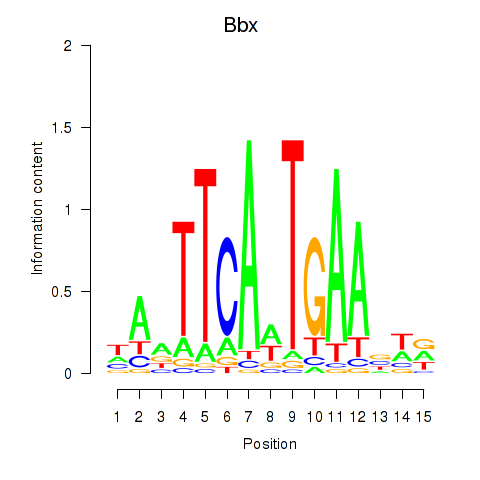
Results for Bbx
Z-value: 0.31
Transcription factors associated with Bbx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bbx
|
ENSRNOG00000001971 | BBX, HMG-box containing |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bbx | rn6_v1_chr11_+_53081025_53081025 | -0.19 | 8.6e-04 | Click! |
Activity profile of Bbx motif
Sorted Z-values of Bbx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_102794168 | 10.80 |
ENSRNOT00000048400
|
AABR07061022.1
|
|
| chr15_+_32614002 | 9.46 |
ENSRNOT00000072962
|
AABR07017902.1
|
|
| chr14_-_5859581 | 8.48 |
ENSRNOT00000052308
|
AABR07014241.1
|
|
| chr10_+_71047310 | 8.25 |
ENSRNOT00000042643
ENSRNOT00000088737 |
Wfdc21
|
WAP four-disulfide core domain 21 |
| chrX_-_110230610 | 7.61 |
ENSRNOT00000093401
|
Serpina7
|
serpin family A member 7 |
| chr14_+_2050483 | 6.26 |
ENSRNOT00000000047
|
Slc26a1
|
solute carrier family 26 member 1 |
| chr11_+_85042348 | 6.07 |
ENSRNOT00000042220
|
AABR07034718.1
|
|
| chr20_-_27782586 | 6.02 |
ENSRNOT00000001093
|
Dse
|
dermatan sulfate epimerase |
| chr16_-_25192675 | 5.92 |
ENSRNOT00000032289
|
March1
|
membrane associated ring-CH-type finger 1 |
| chr17_-_86657473 | 5.24 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr3_-_79282493 | 5.07 |
ENSRNOT00000049832
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr7_-_143497108 | 4.81 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr13_+_92146586 | 4.31 |
ENSRNOT00000004659
|
Olr1588
|
olfactory receptor 1588 |
| chr5_+_151776004 | 3.90 |
ENSRNOT00000009683
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr8_-_97647072 | 3.84 |
ENSRNOT00000030645
|
Tbc1d2b
|
TBC1 domain family, member 2B |
| chr18_+_40596166 | 3.09 |
ENSRNOT00000087984
|
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr13_-_111972603 | 3.02 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr3_-_24601063 | 2.84 |
ENSRNOT00000037043
|
LOC100362366
|
40S ribosomal protein S17-like |
| chr2_+_107233054 | 2.82 |
ENSRNOT00000015573
|
Tbl1xr1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr18_+_72571327 | 2.41 |
ENSRNOT00000089468
|
Smad2
|
SMAD family member 2 |
| chr3_-_173166369 | 1.70 |
ENSRNOT00000090602
|
AABR07054887.1
|
|
| chr1_-_124803363 | 1.68 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr15_-_27140657 | 1.60 |
ENSRNOT00000016727
|
Olr1612
|
olfactory receptor 1612 |
| chr12_-_6630274 | 1.49 |
ENSRNOT00000087840
|
AABR07035194.1
|
|
| chr8_+_61659445 | 1.40 |
ENSRNOT00000023831
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr3_-_43119159 | 1.31 |
ENSRNOT00000041394
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chrX_-_80142902 | 1.19 |
ENSRNOT00000071650
|
Hmgn5
|
high mobility group nucleosome binding domain 5 |
| chr3_-_167225532 | 1.06 |
ENSRNOT00000075012
|
LOC102553249
|
zinc finger protein 120-like |
| chr8_-_94920441 | 0.88 |
ENSRNOT00000014165
|
Cep162
|
centrosomal protein 162 |
| chr1_+_124477164 | 0.73 |
ENSRNOT00000048744
|
Hmgn5b
|
high mobility group nucleosome binding domain 5B |
| chr3_-_173371835 | 0.58 |
ENSRNOT00000073650
|
LOC102557590
|
zinc finger protein 709-like |
| chr4_+_166911595 | 0.56 |
ENSRNOT00000030813
|
Tas2r124
|
taste receptor, type 2, member 124 |
| chr3_-_167270512 | 0.41 |
ENSRNOT00000075542
|
AABR07054708.1
|
|
| chr2_+_240642184 | 0.39 |
ENSRNOT00000083181
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr19_-_37528011 | 0.32 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr3_-_173249556 | 0.28 |
ENSRNOT00000081623
|
AABR07054895.1
|
|
| chr6_-_26135952 | 0.20 |
ENSRNOT00000084751
|
Mrpl33
|
mitochondrial ribosomal protein L33 |
| chrX_-_135419704 | 0.05 |
ENSRNOT00000044435
|
Zfp280c
|
zinc finger protein 280C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Bbx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 1.0 | 3.0 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.7 | 7.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.6 | 2.4 | GO:1900224 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.6 | 11.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.5 | 5.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.4 | 6.3 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.3 | 4.8 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.3 | 1.3 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.2 | 5.9 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 1.7 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 2.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 3.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 3.9 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 3.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 5.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.9 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 5.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 4.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 6.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 1.0 | 3.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.0 | 7.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.6 | 2.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.4 | 6.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.4 | 5.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.3 | 3.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 5.9 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 3.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 4.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 8.3 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 2.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 5.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 5.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.0 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 2.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 5.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 6.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |