Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Batf3
Z-value: 0.56
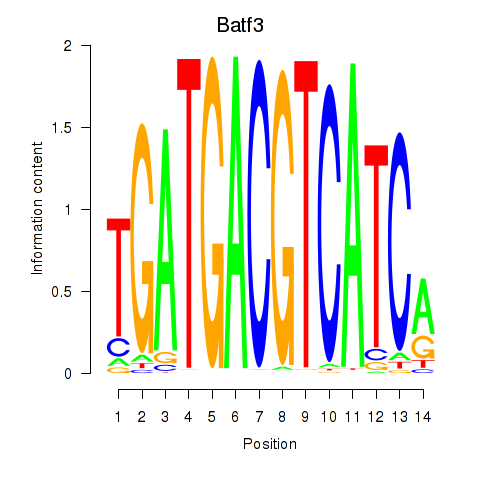
Transcription factors associated with Batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf3
|
ENSRNOG00000003716 | basic leucine zipper ATF-like transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Batf3 | rn6_v1_chr13_+_109713489_109713489 | 0.44 | 7.5e-17 | Click! |
Activity profile of Batf3 motif
Sorted Z-values of Batf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_16970626 | 22.03 |
ENSRNOT00000005383
|
Dusp1
|
dual specificity phosphatase 1 |
| chr10_+_70884531 | 20.18 |
ENSRNOT00000015199
|
Ccl4
|
C-C motif chemokine ligand 4 |
| chr5_+_159845774 | 17.57 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr9_-_26734763 | 17.05 |
ENSRNOT00000082814
|
AABR07067024.1
|
|
| chr2_+_153830586 | 14.98 |
ENSRNOT00000042576
|
Mme
|
membrane metallo-endopeptidase |
| chr4_+_69403735 | 14.84 |
ENSRNOT00000082012
|
Trbv13-1
|
T cell receptor beta, variable 13-1 |
| chr7_+_120580743 | 14.40 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr2_+_189629297 | 13.11 |
ENSRNOT00000049810
|
Dennd4b
|
DENN domain containing 4B |
| chr11_-_62451149 | 12.46 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_+_177495782 | 12.23 |
ENSRNOT00000021020
|
Tead1
|
TEA domain transcription factor 1 |
| chr2_-_66608324 | 11.80 |
ENSRNOT00000077597
|
Cln5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr17_+_10463303 | 11.02 |
ENSRNOT00000060822
|
Rnf44
|
ring finger protein 44 |
| chr15_-_34392066 | 11.00 |
ENSRNOT00000027315
|
Tgm1
|
transglutaminase 1 |
| chr4_-_57625147 | 9.48 |
ENSRNOT00000074649
ENSRNOT00000078961 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chrX_-_45522665 | 9.04 |
ENSRNOT00000030771
|
RGD1562200
|
similar to GS2 gene |
| chr12_+_13102019 | 8.27 |
ENSRNOT00000092628
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr6_+_96171756 | 7.93 |
ENSRNOT00000029328
|
Slc38a6
|
solute carrier family 38, member 6 |
| chr9_+_82370924 | 7.86 |
ENSRNOT00000025219
|
Zfand2b
|
zinc finger AN1-type containing 2B |
| chr2_-_185005572 | 7.41 |
ENSRNOT00000093291
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr3_+_148635775 | 7.36 |
ENSRNOT00000067261
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr4_-_6330699 | 7.33 |
ENSRNOT00000078077
ENSRNOT00000011814 |
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr10_-_88163712 | 7.19 |
ENSRNOT00000005382
ENSRNOT00000084493 |
Krt17
|
keratin 17 |
| chr14_+_104475082 | 6.52 |
ENSRNOT00000084481
|
Rab1a
|
RAB1A, member RAS oncogene family |
| chr5_-_107857320 | 6.08 |
ENSRNOT00000008898
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chr4_+_6330466 | 6.03 |
ENSRNOT00000089582
|
E130116L18Rik
|
RIKEN cDNA E130116L18 gene |
| chr7_-_9834670 | 5.92 |
ENSRNOT00000075919
ENSRNOT00000076994 |
LOC102549817
|
zinc finger protein 124-like |
| chr10_+_11724032 | 5.75 |
ENSRNOT00000008966
|
Trap1
|
TNF receptor-associated protein 1 |
| chr11_-_64752544 | 5.57 |
ENSRNOT00000048738
|
Tmem39a
|
transmembrane protein 39a |
| chr20_+_5262120 | 5.15 |
ENSRNOT00000000535
|
Brd2
|
bromodomain containing 2 |
| chr19_+_56272162 | 5.08 |
ENSRNOT00000030399
|
Afg3l1
|
AFG3(ATPase family gene 3)-like 1 (S. cerevisiae) |
| chr15_+_27739251 | 4.53 |
ENSRNOT00000011840
|
Parp2
|
poly (ADP-ribose) polymerase 2 |
| chr7_+_10962330 | 4.33 |
ENSRNOT00000008360
|
Tle6
|
transducin-like enhancer of split 6 |
| chr14_+_1462358 | 4.25 |
ENSRNOT00000077243
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
| chr2_+_240396152 | 4.17 |
ENSRNOT00000034565
|
Cenpe
|
centromere protein E |
| chr1_-_156296161 | 4.04 |
ENSRNOT00000025653
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr3_-_117389456 | 3.78 |
ENSRNOT00000007103
ENSRNOT00000081533 |
Myef2
|
myelin expression factor 2 |
| chr12_-_2170504 | 3.71 |
ENSRNOT00000001309
|
Xab2
|
XPA binding protein 2 |
| chr20_-_10257044 | 3.65 |
ENSRNOT00000068289
|
Wdr4
|
WD repeat domain 4 |
| chr1_+_201256910 | 3.40 |
ENSRNOT00000090143
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_-_96171629 | 3.38 |
ENSRNOT00000010546
ENSRNOT00000078190 |
Trmt5
|
tRNA methyltransferase 5 |
| chr8_+_114867062 | 3.31 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr4_-_118472179 | 3.09 |
ENSRNOT00000023856
|
Mxd1
|
max dimerization protein 1 |
| chrX_-_158978995 | 3.04 |
ENSRNOT00000001179
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr5_+_148577332 | 2.46 |
ENSRNOT00000016325
|
Snrnp40
|
small nuclear ribonucleoprotein U5 subunit 40 |
| chr4_-_113886994 | 2.45 |
ENSRNOT00000037333
|
Htra2
|
HtrA serine peptidase 2 |
| chr9_-_20219209 | 2.34 |
ENSRNOT00000072086
|
LOC100911548
|
SPRY domain-containing SOCS box protein 2-like |
| chr3_-_120011364 | 2.26 |
ENSRNOT00000018922
|
Fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr1_-_259691742 | 2.22 |
ENSRNOT00000088065
|
Tctn3
|
tectonic family member 3 |
| chr5_-_152464850 | 2.22 |
ENSRNOT00000021937
|
Zfp593
|
zinc finger protein 593 |
| chrX_+_15064594 | 2.22 |
ENSRNOT00000007130
|
Tbc1d25
|
TBC1 domain family, member 25 |
| chr3_-_9236736 | 2.16 |
ENSRNOT00000072628
|
Nup214
|
nucleoporin 214 |
| chr8_+_114866768 | 2.06 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr8_-_128009951 | 2.05 |
ENSRNOT00000018056
|
Oxsr1
|
oxidative-stress responsive 1 |
| chr4_+_157326727 | 1.87 |
ENSRNOT00000020493
|
Spsb2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr9_+_19451630 | 1.77 |
ENSRNOT00000065048
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr9_+_38799881 | 1.75 |
ENSRNOT00000083847
|
AABR07067300.1
|
|
| chr1_-_259106948 | 1.54 |
ENSRNOT00000074429
|
LOC100912537
|
tectonic-3-like |
| chr2_-_152824547 | 1.07 |
ENSRNOT00000019824
|
Dhx36
|
DEAH-box helicase 36 |
| chr4_+_113887115 | 1.03 |
ENSRNOT00000010602
|
Aup1
|
ancient ubiquitous protein 1 |
| chr16_-_69195097 | 0.90 |
ENSRNOT00000018973
|
Erlin2
|
ER lipid raft associated 2 |
| chr12_+_29921443 | 0.61 |
ENSRNOT00000001190
|
Sbds
|
SBDS ribosome assembly guanine nucleotide exchange factor |
| chr1_+_218466289 | 0.41 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr3_+_112371677 | 0.25 |
ENSRNOT00000013316
|
LOC100912076
|
HAUS augmin-like complex subunit 2-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Batf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 17.6 | GO:1901491 | notochord formation(GO:0014028) axial mesoderm morphogenesis(GO:0048319) negative regulation of lymphangiogenesis(GO:1901491) |
| 3.7 | 15.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 3.4 | 20.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 2.0 | 11.8 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 2.0 | 7.9 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 1.9 | 5.7 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 1.9 | 13.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 1.7 | 8.3 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 1.6 | 22.0 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 1.5 | 4.5 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 1.5 | 7.3 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 1.1 | 4.3 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.9 | 12.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.9 | 3.7 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.9 | 18.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.8 | 3.4 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.8 | 4.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.8 | 6.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.7 | 2.0 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.6 | 7.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.5 | 5.1 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.4 | 3.0 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.4 | 2.4 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.4 | 12.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.4 | 6.1 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.4 | 1.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.3 | 9.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.3 | 3.7 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.3 | 0.9 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.3 | 14.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.3 | 2.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.3 | 5.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.2 | 9.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 3.4 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.2 | 2.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 7.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.8 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) bone marrow development(GO:0048539) |
| 0.1 | 5.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 4.0 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 1.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 2.2 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 3.8 | GO:0014902 | myotube differentiation(GO:0014902) |
| 0.0 | 9.1 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 2.2 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.7 | 5.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.7 | 3.7 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.6 | 8.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.5 | 11.8 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.4 | 5.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.3 | 1.0 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 4.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 2.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 2.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 14.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 7.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 11.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 15.0 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 17.6 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.1 | 2.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 6.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 3.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 3.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 22.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 3.4 | 20.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 3.0 | 9.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 1.4 | 4.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.2 | 7.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.1 | 11.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.9 | 8.3 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.9 | 17.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.8 | 3.0 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.7 | 2.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.7 | 3.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.7 | 7.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.6 | 12.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.6 | 3.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.6 | 11.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.5 | 4.5 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.5 | 4.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 6.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 13.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 9.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 2.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 5.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 5.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 20.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 1.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 19.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 2.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 7.9 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 9.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 18.4 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 2.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 5.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 25.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.6 | 22.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 20.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 10.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 4.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 7.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 5.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 6.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 11.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.4 | 12.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 20.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 6.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 7.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 14.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 4.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 9.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |