Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
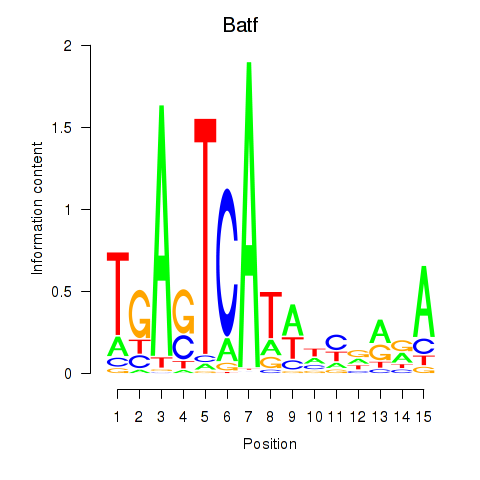
Results for Batf
Z-value: 0.66
Transcription factors associated with Batf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf
|
ENSRNOG00000008588 | basic leucine zipper ATF-like transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Batf | rn6_v1_chr6_+_109562587_109562587 | 0.39 | 5.6e-13 | Click! |
Activity profile of Batf motif
Sorted Z-values of Batf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_227207584 | 26.34 |
ENSRNOT00000065361
ENSRNOT00000080215 |
Myoz2
|
myozenin 2 |
| chr4_-_164051812 | 18.25 |
ENSRNOT00000085719
|
AABR07062183.1
|
|
| chr19_+_37235001 | 16.91 |
ENSRNOT00000020908
|
Nol3
|
nucleolar protein 3 |
| chr4_-_163810403 | 16.34 |
ENSRNOT00000079704
|
AABR07062170.1
|
|
| chr1_+_199555722 | 14.23 |
ENSRNOT00000054983
|
Itgax
|
integrin subunit alpha X |
| chr15_-_28081465 | 13.19 |
ENSRNOT00000033739
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chrX_+_104734082 | 12.35 |
ENSRNOT00000005020
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr8_+_5606592 | 12.19 |
ENSRNOT00000011727
|
Mmp12
|
matrix metallopeptidase 12 |
| chr5_+_64476317 | 11.92 |
ENSRNOT00000017217
|
LOC108348074
|
collagen alpha-1(XV) chain-like |
| chr14_+_34727623 | 11.52 |
ENSRNOT00000071405
ENSRNOT00000090183 |
Kdr
|
kinase insert domain receptor |
| chr16_-_9430743 | 11.25 |
ENSRNOT00000043811
|
Wdfy4
|
WDFY family member 4 |
| chr10_+_69423086 | 10.97 |
ENSRNOT00000000256
|
Ccl7
|
C-C motif chemokine ligand 7 |
| chr3_+_19274273 | 10.85 |
ENSRNOT00000040102
|
AABR07051684.1
|
|
| chr8_-_49038169 | 10.56 |
ENSRNOT00000047303
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr8_+_133210473 | 10.21 |
ENSRNOT00000082774
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr4_+_98481520 | 9.86 |
ENSRNOT00000078381
ENSRNOT00000048493 |
AABR07060886.1
|
|
| chr18_+_56414488 | 9.67 |
ENSRNOT00000088988
|
Csf1r
|
colony stimulating factor 1 receptor |
| chr14_-_81053905 | 9.07 |
ENSRNOT00000045068
ENSRNOT00000040215 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr5_+_154265097 | 9.00 |
ENSRNOT00000012342
|
Cnr2
|
cannabinoid receptor 2 |
| chr10_+_83655460 | 8.91 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr4_-_41212072 | 8.87 |
ENSRNOT00000085596
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr4_-_17440506 | 8.76 |
ENSRNOT00000080931
|
Sema3e
|
semaphorin 3E |
| chr3_-_16441030 | 8.29 |
ENSRNOT00000047784
|
AABR07051532.1
|
|
| chr2_+_153803349 | 8.12 |
ENSRNOT00000088565
|
Mme
|
membrane metallo-endopeptidase |
| chrX_-_31851715 | 7.86 |
ENSRNOT00000068601
|
Vegfd
|
vascular endothelial growth factor D |
| chrX_-_10031167 | 7.47 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chr14_-_28856214 | 7.27 |
ENSRNOT00000044659
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr7_+_145068286 | 6.93 |
ENSRNOT00000088956
ENSRNOT00000065753 |
Nckap1l
|
NCK associated protein 1 like |
| chr3_+_16571602 | 6.65 |
ENSRNOT00000048351
|
LOC100361705
|
rCG64259-like |
| chrX_+_43625169 | 5.64 |
ENSRNOT00000086311
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr3_-_48535909 | 5.39 |
ENSRNOT00000008148
|
Fap
|
fibroblast activation protein, alpha |
| chr17_-_22143324 | 5.30 |
ENSRNOT00000019361
|
Edn1
|
endothelin 1 |
| chr4_-_155690869 | 5.28 |
ENSRNOT00000012216
|
C3ar1
|
complement C3a receptor 1 |
| chr4_-_103761881 | 5.28 |
ENSRNOT00000084103
|
AABR07061087.1
|
|
| chr2_-_32518643 | 4.85 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_118547190 | 4.71 |
ENSRNOT00000083676
ENSRNOT00000090301 ENSRNOT00000013410 |
Kcnmb2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr4_+_3043231 | 4.59 |
ENSRNOT00000013732
|
Il6
|
interleukin 6 |
| chr13_-_69984785 | 4.54 |
ENSRNOT00000083995
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr20_+_18833481 | 3.86 |
ENSRNOT00000080846
|
Bicc1
|
BicC family RNA binding protein 1 |
| chr18_+_15467870 | 3.81 |
ENSRNOT00000091696
|
B4galt6
|
beta-1,4-galactosyltransferase 6 |
| chr6_+_136358057 | 3.62 |
ENSRNOT00000092741
|
Klc1
|
kinesin light chain 1 |
| chr10_-_37455022 | 3.55 |
ENSRNOT00000016742
|
LOC103694902
|
ubiquitin-conjugating enzyme E2 B |
| chr3_-_29984201 | 3.21 |
ENSRNOT00000006350
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr10_-_83655182 | 3.18 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr5_-_163231494 | 3.16 |
ENSRNOT00000085929
ENSRNOT00000022968 |
Tnfrsf8
|
TNF receptor superfamily member 8 |
| chr2_-_177950517 | 3.09 |
ENSRNOT00000012792
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr14_-_85484275 | 2.98 |
ENSRNOT00000083770
|
Kremen1
|
kringle containing transmembrane protein 1 |
| chr9_+_117737611 | 2.97 |
ENSRNOT00000022396
|
Zfp161
|
zinc finger protein 161 |
| chr4_-_138830117 | 2.97 |
ENSRNOT00000008403
|
Il5ra
|
interleukin 5 receptor subunit alpha |
| chr2_-_123972356 | 2.86 |
ENSRNOT00000023348
|
Il21
|
interleukin 21 |
| chr2_+_144861455 | 2.71 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr2_+_2605996 | 2.70 |
ENSRNOT00000016372
|
Glrx
|
glutaredoxin |
| chr18_+_3861539 | 2.70 |
ENSRNOT00000015363
|
Lama3
|
laminin subunit alpha 3 |
| chr1_+_220428481 | 2.57 |
ENSRNOT00000027335
|
LOC108348044
|
ras and Rab interactor 1 |
| chr1_+_220335254 | 2.53 |
ENSRNOT00000072261
|
Rin1
|
Ras and Rab interactor 1 |
| chr15_-_41508952 | 2.51 |
ENSRNOT00000064503
|
AABR07018124.1
|
|
| chr4_-_22307453 | 2.32 |
ENSRNOT00000047126
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr3_-_161812243 | 2.26 |
ENSRNOT00000025238
ENSRNOT00000087646 |
Slc35c2
|
solute carrier family 35 member C2 |
| chrX_-_111179152 | 2.23 |
ENSRNOT00000089115
|
Morc4
|
MORC family CW-type zinc finger 4 |
| chr13_-_90977734 | 2.17 |
ENSRNOT00000011869
|
Slamf8
|
SLAM family member 8 |
| chr3_+_2642531 | 2.10 |
ENSRNOT00000081798
|
Fut7
|
fucosyltransferase 7 |
| chr1_-_66645214 | 1.93 |
ENSRNOT00000084168
|
LOC100365062
|
eukaryotic translation initiation factor 3, subunit 6 48kDa-like |
| chr5_-_77031120 | 1.92 |
ENSRNOT00000022987
|
Inip
|
INTS3 and NABP interacting protein |
| chr1_-_66639205 | 1.92 |
ENSRNOT00000082190
|
LOC100909481
|
eukaryotic translation initiation factor 3 subunit E-like |
| chr1_+_46230557 | 1.70 |
ENSRNOT00000092849
|
Arid1b
|
AT-rich interaction domain 1B |
| chr16_-_19637609 | 1.65 |
ENSRNOT00000021742
|
Zfp709
|
zinc finger protein 709 |
| chr14_-_24080470 | 1.64 |
ENSRNOT00000002745
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr10_-_85084850 | 1.59 |
ENSRNOT00000012462
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr6_+_135610743 | 1.53 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr8_+_85951191 | 1.48 |
ENSRNOT00000037665
|
Cd109
|
CD109 molecule |
| chr18_+_36596585 | 1.41 |
ENSRNOT00000036613
|
Rbm27
|
RNA binding motif protein 27 |
| chr3_+_102703487 | 1.38 |
ENSRNOT00000089603
|
AC121203.2
|
|
| chr3_-_78640261 | 1.26 |
ENSRNOT00000048787
|
Olr717
|
olfactory receptor 717 |
| chr2_-_251970768 | 1.22 |
ENSRNOT00000020141
|
Wdr63
|
WD repeat domain 63 |
| chr3_-_64095120 | 1.16 |
ENSRNOT00000016837
|
Sestd1
|
SEC14 and spectrin domain containing 1 |
| chr12_-_21746236 | 1.16 |
ENSRNOT00000001869
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr5_+_169181418 | 1.14 |
ENSRNOT00000004508
|
Klhl21
|
kelch-like family member 21 |
| chr2_+_149843282 | 1.01 |
ENSRNOT00000074805
|
RGD1561998
|
similar to hypothetical protein C130079G13 |
| chr8_+_19886382 | 0.95 |
ENSRNOT00000034254
|
Zfp317
|
zinc finger protein 317 |
| chr18_+_86116044 | 0.95 |
ENSRNOT00000058160
ENSRNOT00000076159 |
Rttn
|
rotatin |
| chr1_+_72210037 | 0.90 |
ENSRNOT00000058999
|
Vom1r37
|
vomeronasal 1 receptor 37 |
| chrM_+_7919 | 0.86 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr2_-_183210799 | 0.79 |
ENSRNOT00000085382
|
Trim2
|
tripartite motif-containing 2 |
| chr4_-_72023952 | 0.79 |
ENSRNOT00000059395
|
Fam115e
|
family with sequence similarity 115, member E |
| chr7_+_59200918 | 0.70 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr3_-_20883265 | 0.68 |
ENSRNOT00000052047
|
Olr414
|
olfactory receptor 414 |
| chr1_-_98486976 | 0.66 |
ENSRNOT00000024083
|
Etfb
|
electron transfer flavoprotein beta subunit |
| chr8_+_114866768 | 0.60 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr2_+_243425007 | 0.43 |
ENSRNOT00000082894
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr3_-_78555530 | 0.29 |
ENSRNOT00000089672
|
Olr711
|
olfactory receptor 711 |
| chr1_+_199323628 | 0.25 |
ENSRNOT00000036187
|
Zfp646
|
zinc finger protein 646 |
| chr3_+_75465399 | 0.09 |
ENSRNOT00000057479
|
Olr562
|
olfactory receptor 562 |
| chr2_+_46210713 | 0.09 |
ENSRNOT00000074742
|
LOC100910111
|
olfactory receptor 147-like |
| chr3_-_78409263 | 0.03 |
ENSRNOT00000041001
|
Olr704
|
olfactory receptor 704 |
| chrX_+_908044 | 0.01 |
ENSRNOT00000072087
|
Zfp300
|
zinc finger protein 300 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Batf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.9 | GO:0014736 | negative regulation of muscle atrophy(GO:0014736) |
| 3.4 | 10.2 | GO:2000464 | positive regulation of astrocyte chemotaxis(GO:2000464) |
| 3.0 | 12.2 | GO:0072641 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 2.3 | 6.9 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 2.0 | 8.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 1.9 | 9.7 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 1.9 | 11.5 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 1.8 | 11.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.8 | 5.4 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 1.8 | 5.3 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 1.8 | 5.3 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 1.6 | 7.9 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 1.5 | 12.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 1.5 | 4.6 | GO:2000660 | negative regulation of chemokine biosynthetic process(GO:0045079) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.5 | 9.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 1.4 | 5.6 | GO:0032919 | spermine acetylation(GO:0032919) |
| 1.1 | 3.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 1.1 | 3.2 | GO:0032639 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 1.0 | 9.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.9 | 2.7 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.8 | 10.6 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.8 | 3.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.8 | 2.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.7 | 3.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.6 | 2.3 | GO:1905235 | carbohydrate export(GO:0033231) cellular response to mycotoxin(GO:0036146) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.5 | 2.2 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of neutrophil migration(GO:1902623) |
| 0.5 | 2.9 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.4 | 1.7 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.4 | 26.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.4 | 7.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.3 | 4.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 3.8 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.3 | 8.8 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.2 | 1.5 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.2 | 14.2 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.2 | 2.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 3.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 1.2 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.2 | 2.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) fucosylation(GO:0036065) |
| 0.2 | 0.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 1.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 2.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 3.0 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.1 | 1.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 13.2 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 2.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 6.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 3.0 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.6 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 1.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.7 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 1.0 | GO:0034605 | cellular response to heat(GO:0034605) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.5 | 4.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 1.4 | 11.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.9 | 5.3 | GO:0033093 | Weibel-Palade body(GO:0033093) rough endoplasmic reticulum lumen(GO:0048237) |
| 0.8 | 6.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.7 | 10.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 2.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.5 | 3.8 | GO:0033290 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.5 | 1.9 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 2.7 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.4 | 14.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 3.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 9.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 16.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.2 | 8.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 26.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 1.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 7.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 9.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 5.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 3.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 4.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 8.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 14.4 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 2.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 8.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 5.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 19.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 6.3 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.5 | GO:0030425 | dendrite(GO:0030425) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 26.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 4.2 | 16.9 | GO:0035877 | death effector domain binding(GO:0035877) |
| 3.5 | 21.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 2.3 | 9.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 1.9 | 5.6 | GO:0019809 | spermidine binding(GO:0019809) |
| 1.8 | 5.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.4 | 11.5 | GO:0038085 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 1.0 | 3.8 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.9 | 4.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.9 | 7.9 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.7 | 8.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.7 | 4.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.6 | 2.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.5 | 5.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.5 | 3.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.5 | 5.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.5 | 2.3 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.4 | 2.7 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.4 | 2.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 3.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 9.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 9.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.2 | 4.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 16.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 13.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 3.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 13.8 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 0.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 6.1 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 1.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 3.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 3.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 3.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 7.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 4.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 4.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 5.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 11.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 11.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.5 | 14.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.4 | 3.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 4.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.3 | 8.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 8.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 8.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 9.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 5.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 3.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 2.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 10.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 3.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 5.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 1.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 8.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 8.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 8.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 10.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.0 | 11.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.5 | 8.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 8.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.4 | 4.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 5.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 11.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 3.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 4.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 14.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 3.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 13.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 5.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |