Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
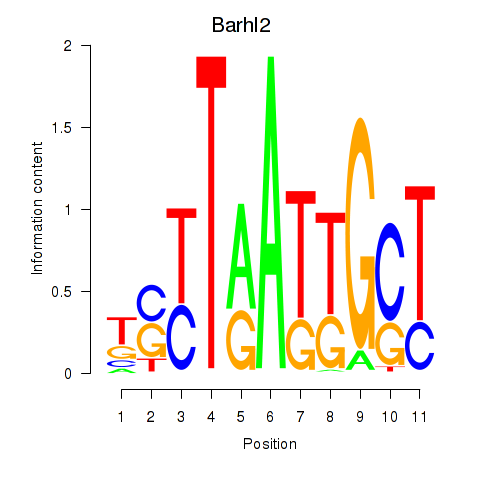
Results for Barhl2
Z-value: 0.61
Transcription factors associated with Barhl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Barhl2
|
ENSRNOG00000002117 | BarH-like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barhl2 | rn6_v1_chr14_+_4362717_4362717 | -0.32 | 7.5e-09 | Click! |
Activity profile of Barhl2 motif
Sorted Z-values of Barhl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_88122233 | 26.13 |
ENSRNOT00000083895
ENSRNOT00000005285 |
Krt14
|
keratin 14 |
| chr17_-_9762813 | 20.43 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr20_-_4070721 | 19.39 |
ENSRNOT00000000523
|
RT1-Ba
|
RT1 class II, locus Ba |
| chr10_+_31324512 | 16.07 |
ENSRNOT00000008559
|
Fndc9
|
fibronectin type III domain containing 9 |
| chr7_+_141249044 | 15.97 |
ENSRNOT00000084911
|
Aqp5
|
aquaporin 5 |
| chr3_+_117421604 | 14.47 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr13_+_47572219 | 13.41 |
ENSRNOT00000088449
ENSRNOT00000087664 ENSRNOT00000005853 |
Pigr
|
polymeric immunoglobulin receptor |
| chr5_-_159946446 | 13.35 |
ENSRNOT00000089184
|
Clcnka
|
chloride voltage-gated channel Ka |
| chr1_+_81230612 | 12.81 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr17_-_43807540 | 12.40 |
ENSRNOT00000074763
|
LOC684762
|
similar to CG31613-PA |
| chr12_-_22138382 | 11.06 |
ENSRNOT00000001899
|
Lrch4
|
leucine rich repeats and calponin homology domain containing 4 |
| chr5_-_159962218 | 10.96 |
ENSRNOT00000050729
|
Clcnkb
|
chloride voltage-gated channel Kb |
| chr6_-_92121351 | 10.48 |
ENSRNOT00000006374
|
Cdkl1
|
cyclin dependent kinase like 1 |
| chr6_-_131914028 | 8.60 |
ENSRNOT00000007602
|
Bcl11b
|
B-cell CLL/lymphoma 11B |
| chr7_-_143228060 | 8.50 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr20_-_157665 | 8.06 |
ENSRNOT00000048858
ENSRNOT00000079494 |
RT1-CE10
|
RT1 class I, locus CE10 |
| chr1_+_193187972 | 7.84 |
ENSRNOT00000090172
ENSRNOT00000065342 |
Slc5a11
|
solute carrier family 5 member 11 |
| chr14_-_21299068 | 7.72 |
ENSRNOT00000065778
|
Amtn
|
amelotin |
| chr20_+_44680449 | 7.28 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr12_-_51877624 | 6.90 |
ENSRNOT00000056800
|
Chek2
|
checkpoint kinase 2 |
| chr7_-_142062870 | 6.80 |
ENSRNOT00000026531
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr20_-_157861 | 6.78 |
ENSRNOT00000084461
|
RT1-CE10
|
RT1 class I, locus CE10 |
| chr10_-_4910305 | 6.60 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr4_+_158088505 | 6.42 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr20_+_4106189 | 6.25 |
ENSRNOT00000042571
|
RT1-Db2
|
RT1 class II, locus Db2 |
| chr1_-_175586802 | 6.13 |
ENSRNOT00000071333
|
AABR07005027.1
|
|
| chr3_-_165700489 | 6.07 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr10_-_86932154 | 5.92 |
ENSRNOT00000085344
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr2_-_112831476 | 5.83 |
ENSRNOT00000018055
|
Ect2
|
epithelial cell transforming 2 |
| chrX_+_71335491 | 5.73 |
ENSRNOT00000076003
|
Nono
|
non-POU domain containing, octamer-binding |
| chr4_+_57378069 | 5.65 |
ENSRNOT00000080173
ENSRNOT00000011851 |
Nrf1
|
nuclear respiratory factor 1 |
| chr5_-_115387377 | 5.53 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr15_+_31178339 | 5.39 |
ENSRNOT00000089064
|
AABR07017798.2
|
|
| chr16_-_49453394 | 5.28 |
ENSRNOT00000041617
|
Lrp2bp
|
Lrp2 binding protein |
| chr10_+_105393072 | 5.25 |
ENSRNOT00000013359
|
Ubald2
|
UBA-like domain containing 2 |
| chr1_-_89539210 | 5.15 |
ENSRNOT00000077462
ENSRNOT00000028644 |
Hpn
|
hepsin |
| chr10_+_59799123 | 5.04 |
ENSRNOT00000026493
|
Trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr13_-_83425641 | 4.95 |
ENSRNOT00000063870
|
Tbx19
|
T-box 19 |
| chr8_+_48665652 | 4.72 |
ENSRNOT00000059715
|
H2afx
|
H2A histone family, member X |
| chr15_+_29157294 | 4.63 |
ENSRNOT00000085162
|
AABR07017604.1
|
|
| chr1_-_247821728 | 4.57 |
ENSRNOT00000021958
|
Ermp1
|
endoplasmic reticulum metallopeptidase 1 |
| chr10_+_104523996 | 4.43 |
ENSRNOT00000065339
ENSRNOT00000086747 |
Itgb4
|
integrin subunit beta 4 |
| chr9_+_15513063 | 4.16 |
ENSRNOT00000020499
|
Taf8
|
TATA-box binding protein associated factor 8 |
| chr13_+_25966810 | 4.10 |
ENSRNOT00000086167
|
Zcchc2
|
zinc finger CCHC-type containing 2 |
| chr7_+_142397371 | 4.10 |
ENSRNOT00000040890
ENSRNOT00000065379 |
Slc4a8
|
solute carrier family 4 member 8 |
| chr7_-_142063212 | 3.97 |
ENSRNOT00000089912
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr1_+_178351680 | 3.93 |
ENSRNOT00000018532
ENSRNOT00000076925 ENSRNOT00000076430 ENSRNOT00000076756 |
Far1
|
fatty acyl CoA reductase 1 |
| chr1_-_64090017 | 3.57 |
ENSRNOT00000086622
ENSRNOT00000091654 |
Rps9
|
ribosomal protein S9 |
| chr12_-_24556976 | 3.57 |
ENSRNOT00000041798
|
Bcl7b
|
BCL tumor suppressor 7B |
| chr1_-_64021321 | 3.39 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chr10_-_15098791 | 3.14 |
ENSRNOT00000026139
|
Chtf18
|
chromosome transmission fidelity factor 18 |
| chr9_+_92565366 | 3.00 |
ENSRNOT00000023184
|
Csprs
|
component of Sp100-rs |
| chr7_-_73130740 | 2.96 |
ENSRNOT00000075584
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr10_-_48038647 | 2.80 |
ENSRNOT00000078448
|
Prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr17_-_84830185 | 2.68 |
ENSRNOT00000040697
|
Skida1
|
SKI/DACH domain containing 1 |
| chr17_+_17965881 | 2.66 |
ENSRNOT00000021751
|
Dek
|
DEK proto-oncogene |
| chr3_-_171342646 | 2.62 |
ENSRNOT00000071853
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_-_128149220 | 2.46 |
ENSRNOT00000014204
|
Gsc
|
goosecoid homeobox |
| chr13_+_25966428 | 2.45 |
ENSRNOT00000061814
ENSRNOT00000003870 |
Zcchc2
|
zinc finger CCHC-type containing 2 |
| chr3_+_176821652 | 2.43 |
ENSRNOT00000055030
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr3_-_147487170 | 2.39 |
ENSRNOT00000077580
|
Fam110a
|
family with sequence similarity 110, member A |
| chrX_-_115496045 | 2.33 |
ENSRNOT00000051134
|
LOC100361854
|
ribosomal protein S26-like |
| chr2_+_210880777 | 2.32 |
ENSRNOT00000026237
|
Gnat2
|
G protein subunit alpha transducin 2 |
| chr15_+_44441856 | 2.26 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr5_+_144106802 | 2.23 |
ENSRNOT00000035637
ENSRNOT00000079779 |
Lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr6_+_3375476 | 2.22 |
ENSRNOT00000045349
|
LOC102551744
|
eukaryotic translation initiation factor 1-like |
| chr5_-_164679654 | 2.21 |
ENSRNOT00000035695
|
LOC100911456
|
migration and invasion-inhibitory protein-like |
| chr2_-_197935567 | 2.18 |
ENSRNOT00000085404
|
Rprd2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr1_-_137359072 | 2.12 |
ENSRNOT00000014553
|
Klhl25
|
kelch-like family member 25 |
| chr1_-_164101578 | 2.06 |
ENSRNOT00000022176
|
Uvrag
|
UV radiation resistance associated |
| chr10_-_12030416 | 2.03 |
ENSRNOT00000010442
|
Zfp263
|
zinc finger protein 263 |
| chr15_-_83494423 | 2.02 |
ENSRNOT00000037588
|
Dis3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr10_-_83655182 | 1.97 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chrX_+_984798 | 1.96 |
ENSRNOT00000073016
|
Zfp182
|
zinc finger protein 182 |
| chr11_+_82194657 | 1.95 |
ENSRNOT00000002445
|
Etv5
|
ets variant 5 |
| chr16_+_6903212 | 1.93 |
ENSRNOT00000023314
|
Tmem110
|
transmembrane protein 110 |
| chr4_-_66955732 | 1.89 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr10_-_104482838 | 1.88 |
ENSRNOT00000007246
|
Recql5
|
RecQ like helicase 5 |
| chr14_+_43512901 | 1.87 |
ENSRNOT00000050664
|
Apbb2
|
amyloid beta precursor protein binding family B member 2 |
| chr17_-_42031265 | 1.87 |
ENSRNOT00000068021
|
Dcdc2
|
doublecortin domain containing 2 |
| chr17_+_78735598 | 1.87 |
ENSRNOT00000020854
|
Hspa14
|
heat shock protein family A, member 14 |
| chr5_-_57845509 | 1.87 |
ENSRNOT00000035541
|
Kif24
|
kinesin family member 24 |
| chr16_-_75855745 | 1.82 |
ENSRNOT00000031291
|
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr16_+_71787966 | 1.80 |
ENSRNOT00000080084
|
Htra4
|
HtrA serine peptidase 4 |
| chr11_+_73930753 | 1.71 |
ENSRNOT00000087747
|
RGD1562415
|
similar to 40S ribosomal protein S26 |
| chr20_+_27975549 | 1.67 |
ENSRNOT00000092075
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr1_-_220067123 | 1.61 |
ENSRNOT00000071020
ENSRNOT00000072373 |
Rbm14
|
RNA binding motif protein 14 |
| chr9_+_3238077 | 1.60 |
ENSRNOT00000088083
|
RGD1559808
|
similar to 40S ribosomal protein S26 |
| chr18_+_56364620 | 1.54 |
ENSRNOT00000068535
ENSRNOT00000086033 |
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr13_-_111972603 | 1.53 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr9_+_81644355 | 1.46 |
ENSRNOT00000071700
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chr1_-_64147251 | 1.41 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr2_+_197720259 | 1.33 |
ENSRNOT00000070919
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr3_+_45683993 | 1.19 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr1_-_64162461 | 1.13 |
ENSRNOT00000091739
|
Prpf31
|
pre-mRNA processing factor 31 |
| chr5_+_150957001 | 0.90 |
ENSRNOT00000017549
|
Rpa2
|
replication protein A2 |
| chr20_-_23467190 | 0.78 |
ENSRNOT00000047773
|
RGD1566373
|
similar to large subunit ribosomal protein L36a |
| chr3_+_147422095 | 0.73 |
ENSRNOT00000006786
|
Angpt4
|
angiopoietin 4 |
| chr6_-_36876805 | 0.69 |
ENSRNOT00000006482
|
Msgn1
|
mesogenin 1 |
| chr20_+_4369178 | 0.55 |
ENSRNOT00000088079
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr13_-_68360664 | 0.55 |
ENSRNOT00000030971
|
Hmcn1
|
hemicentin 1 |
| chr10_+_104483042 | 0.52 |
ENSRNOT00000007362
|
Sap30bp
|
SAP30 binding protein |
| chr8_-_12355091 | 0.49 |
ENSRNOT00000009318
|
Cep57
|
centrosomal protein 57 |
| chrX_+_40460047 | 0.48 |
ENSRNOT00000010970
|
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr12_+_12859661 | 0.43 |
ENSRNOT00000001404
|
Usp42
|
ubiquitin specific peptidase 42 |
| chrX_+_110789269 | 0.31 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr4_+_121912352 | 0.24 |
ENSRNOT00000086882
|
Vom1r98
|
vomeronasal 1 receptor 98 |
| chr12_+_38965063 | 0.19 |
ENSRNOT00000074936
|
Morn3
|
MORN repeat containing 3 |
| chr9_+_40817654 | 0.17 |
ENSRNOT00000037392
|
AABR07067339.1
|
|
| chrX_-_25628272 | 0.10 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chrX_+_50599060 | 0.08 |
ENSRNOT00000058420
|
AABR07038477.1
|
|
| chr10_-_44095775 | 0.06 |
ENSRNOT00000047892
|
Olr1422
|
olfactory receptor 1422 |
| chr1_+_64162672 | 0.06 |
ENSRNOT00000089713
|
Tfpt
|
TCF3 (E2A) fusion partner |
| chr6_-_115616766 | 0.06 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr1_-_185143272 | 0.03 |
ENSRNOT00000027752
|
Nucb2
|
nucleobindin 2 |
| chr1_-_148847662 | 0.01 |
ENSRNOT00000090329
|
LOC100911376
|
olfactory receptor 226-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Barhl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.4 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 4.8 | 14.5 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 4.5 | 13.4 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 4.4 | 26.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 3.6 | 10.8 | GO:0015676 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) |
| 3.3 | 13.3 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 2.6 | 7.7 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 2.3 | 16.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.7 | 7.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 1.7 | 6.9 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 1.7 | 5.2 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 1.7 | 5.0 | GO:0090210 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) regulation of establishment of blood-brain barrier(GO:0090210) |
| 1.4 | 8.6 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 1.4 | 11.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 1.2 | 12.8 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 1.0 | 19.4 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 1.0 | 5.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.9 | 11.1 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.8 | 6.4 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.7 | 2.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.6 | 5.7 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.6 | 2.4 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.6 | 6.3 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.6 | 3.9 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.5 | 1.5 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.5 | 4.4 | GO:0061450 | nail development(GO:0035878) trophoblast cell migration(GO:0061450) |
| 0.4 | 2.6 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.4 | 2.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.4 | 2.0 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.4 | 1.5 | GO:0072275 | cell migration involved in vasculogenesis(GO:0035441) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.4 | 7.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.4 | 1.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.4 | 2.3 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.4 | 1.9 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.4 | 5.8 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 3.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 14.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.3 | 2.7 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.3 | 4.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.2 | 2.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 7.8 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 1.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 1.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.2 | 5.7 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.2 | 1.7 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 2.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 1.9 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 0.5 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 1.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 4.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 4.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 12.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 6.6 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.1 | 4.9 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 2.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 1.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 2.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 4.6 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 1.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.7 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 1.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 2.1 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 5.3 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 2.8 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.3 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 1.6 | 25.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.5 | 5.9 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.5 | 5.8 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.1 | 6.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.9 | 14.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.8 | 5.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.6 | 2.3 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.6 | 2.8 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.5 | 42.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.4 | 24.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.3 | 3.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.3 | 5.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 2.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.3 | 1.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 13.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 7.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 20.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 4.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 2.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 18.3 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 12.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 1.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 4.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 12.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 41.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 4.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 4.1 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 2.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 3.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 2.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 6.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.7 | GO:0005840 | ribosome(GO:0005840) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 26.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 4.8 | 14.5 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 4.1 | 20.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 3.6 | 10.8 | GO:0015100 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) nickel cation transmembrane transporter activity(GO:0015099) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) chromium ion transmembrane transporter activity(GO:0070835) |
| 1.8 | 12.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.7 | 5.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 1.5 | 5.9 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.4 | 7.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 1.3 | 24.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 1.1 | 16.0 | GO:0015250 | water channel activity(GO:0015250) |
| 1.1 | 13.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.9 | 4.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.8 | 34.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.8 | 7.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.7 | 2.2 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.5 | 1.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.5 | 1.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.5 | 1.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.4 | 5.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.4 | 10.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.4 | 6.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.4 | 2.8 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.3 | 3.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 1.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.3 | 1.1 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 4.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.7 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 2.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 19.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 10.7 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 2.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 2.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 2.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 5.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 5.7 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 8.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 5.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 7.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 32.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 5.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 15.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 10.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 11.6 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 4.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 13.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 5.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 5.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 2.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 7.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 16.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.6 | 6.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 6.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 39.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 5.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 2.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 2.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 3.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 2.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 7.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 5.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 4.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 2.3 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 5.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 4.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 3.3 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 1.5 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |