Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Barhl1
Z-value: 0.55
Transcription factors associated with Barhl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Barhl1
|
ENSRNOG00000013209 | BarH-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barhl1 | rn6_v1_chr3_-_7498555_7498555 | -0.05 | 4.2e-01 | Click! |
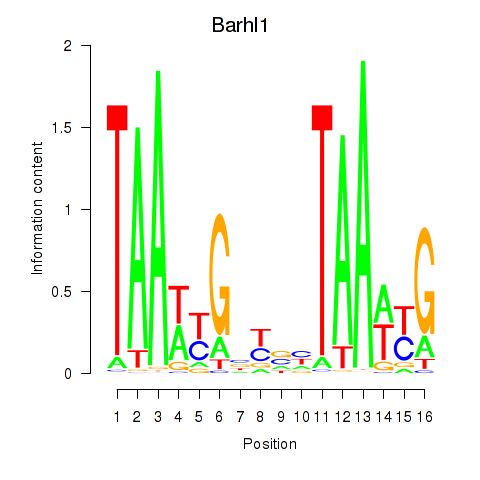
Activity profile of Barhl1 motif
Sorted Z-values of Barhl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_5606592 | 16.87 |
ENSRNOT00000011727
|
Mmp12
|
matrix metallopeptidase 12 |
| chr1_+_279896973 | 14.40 |
ENSRNOT00000068119
|
Pnliprp2
|
pancreatic lipase related protein 2 |
| chr6_-_138852571 | 11.67 |
ENSRNOT00000081803
|
AABR07065656.8
|
|
| chr6_-_139747737 | 11.66 |
ENSRNOT00000090626
|
AABR07065705.3
|
|
| chrX_-_118459355 | 10.82 |
ENSRNOT00000084465
|
Il13ra2
|
interleukin 13 receptor subunit alpha 2 |
| chr4_-_164406146 | 10.53 |
ENSRNOT00000090110
|
Klra22
|
killer cell lectin-like receptor subfamily A, member 22 |
| chr6_-_139041812 | 10.30 |
ENSRNOT00000074510
|
AABR07065656.1
|
|
| chr6_+_139523337 | 9.53 |
ENSRNOT00000090711
|
AABR07065699.4
|
|
| chr11_-_69201380 | 9.48 |
ENSRNOT00000085618
|
Mylk
|
myosin light chain kinase |
| chrX_+_14578264 | 9.18 |
ENSRNOT00000038994
|
Cybb
|
cytochrome b-245 beta chain |
| chr2_-_157759819 | 8.95 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr14_+_59611434 | 8.35 |
ENSRNOT00000065366
|
Cckar
|
cholecystokinin A receptor |
| chr4_-_70934295 | 7.77 |
ENSRNOT00000020616
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr3_+_16571602 | 7.13 |
ENSRNOT00000048351
|
LOC100361705
|
rCG64259-like |
| chr3_+_94549180 | 6.90 |
ENSRNOT00000016318
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr4_-_165026414 | 6.80 |
ENSRNOT00000071421
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr3_+_103753238 | 6.67 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr2_+_185846232 | 6.45 |
ENSRNOT00000023418
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr3_-_112985318 | 6.32 |
ENSRNOT00000015556
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr18_+_56431820 | 6.08 |
ENSRNOT00000079360
ENSRNOT00000049357 |
Csf1r
|
colony stimulating factor 1 receptor |
| chr2_-_49128501 | 5.76 |
ENSRNOT00000073847
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr14_+_17210733 | 5.74 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr13_+_47572219 | 5.43 |
ENSRNOT00000088449
ENSRNOT00000087664 ENSRNOT00000005853 |
Pigr
|
polymeric immunoglobulin receptor |
| chr16_-_3765917 | 5.32 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr1_-_73732118 | 4.88 |
ENSRNOT00000077964
|
Leng8
|
leukocyte receptor cluster member 8 |
| chr9_+_71915421 | 4.64 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr3_+_114102875 | 4.38 |
ENSRNOT00000023209
|
Trim69
|
tripartite motif-containing 69 |
| chr1_-_37932084 | 4.34 |
ENSRNOT00000022527
|
LOC102547287
|
zinc finger protein 728-like |
| chr15_+_31417147 | 4.32 |
ENSRNOT00000092182
|
AABR07017824.2
|
|
| chr16_+_22979444 | 4.15 |
ENSRNOT00000017822
|
Csgalnact1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr10_+_90376933 | 4.15 |
ENSRNOT00000028557
|
Grn
|
granulin precursor |
| chr8_-_104155775 | 4.00 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr10_+_90377103 | 3.50 |
ENSRNOT00000040472
|
Grn
|
granulin precursor |
| chr7_+_130308532 | 3.39 |
ENSRNOT00000011941
|
Miox
|
myo-inositol oxygenase |
| chr4_+_57378069 | 3.38 |
ENSRNOT00000080173
ENSRNOT00000011851 |
Nrf1
|
nuclear respiratory factor 1 |
| chr1_+_53220397 | 3.36 |
ENSRNOT00000089989
|
AABR07001573.1
|
|
| chr15_-_44673765 | 3.35 |
ENSRNOT00000048783
|
AABR07018167.1
|
|
| chr20_+_3992496 | 3.23 |
ENSRNOT00000082047
|
Psmb8
|
proteasome subunit beta 8 |
| chr7_+_76092503 | 3.20 |
ENSRNOT00000079692
|
Grhl2
|
grainyhead-like transcription factor 2 |
| chr5_+_144927759 | 3.20 |
ENSRNOT00000051387
|
AABR07049918.1
|
|
| chr3_+_5709236 | 3.12 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr15_+_30570483 | 2.93 |
ENSRNOT00000071740
|
AABR07017745.1
|
|
| chr3_+_11207542 | 2.92 |
ENSRNOT00000013546
|
Prrc2b
|
proline-rich coiled-coil 2B |
| chr20_-_32296840 | 2.80 |
ENSRNOT00000080727
|
Stox1
|
storkhead box 1 |
| chr12_+_12859661 | 2.73 |
ENSRNOT00000001404
|
Usp42
|
ubiquitin specific peptidase 42 |
| chr11_-_45124423 | 2.67 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr3_-_59166356 | 2.66 |
ENSRNOT00000047713
|
AABR07052519.1
|
|
| chr7_+_144080614 | 2.56 |
ENSRNOT00000077687
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr15_+_31010891 | 2.43 |
ENSRNOT00000088134
|
AABR07017781.1
|
|
| chr1_-_73226777 | 2.36 |
ENSRNOT00000025080
|
Ncr1
|
natural cytotoxicity triggering receptor 1 |
| chr2_-_30340103 | 2.14 |
ENSRNOT00000024023
ENSRNOT00000067722 |
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr6_-_75670135 | 2.05 |
ENSRNOT00000007300
|
Snx6
|
sorting nexin 6 |
| chr6_-_50941248 | 1.85 |
ENSRNOT00000084533
|
Dus4l
|
dihydrouridine synthase 4-like |
| chr4_-_85174931 | 1.84 |
ENSRNOT00000014324
ENSRNOT00000088085 |
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr1_-_197801634 | 1.74 |
ENSRNOT00000090200
|
Nfatc2ip
|
nuclear factor of activated T-cells 2 interacting protein |
| chr4_+_157864969 | 1.73 |
ENSRNOT00000048529
|
Tnfrsf1a
|
TNF receptor superfamily member 1A |
| chr10_+_10530365 | 1.66 |
ENSRNOT00000003843
|
Eef2kmt
|
eukaryotic elongation factor 2 lysine methyltransferase |
| chr18_+_4325875 | 1.65 |
ENSRNOT00000073771
|
Impact
|
impact RWD domain protein |
| chr1_+_205911552 | 1.54 |
ENSRNOT00000024751
|
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
| chr1_-_8038665 | 1.45 |
ENSRNOT00000046539
|
Aig1
|
androgen-induced 1 |
| chr19_-_25955371 | 1.43 |
ENSRNOT00000004042
ENSRNOT00000084123 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chrX_+_123751089 | 1.41 |
ENSRNOT00000092384
|
Nkap
|
NFKB activating protein |
| chr10_-_90127600 | 1.28 |
ENSRNOT00000028368
|
Lsm12
|
LSM12 homolog |
| chr3_+_154437571 | 1.24 |
ENSRNOT00000019331
|
AABR07054456.1
|
|
| chr2_-_170301348 | 1.16 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr9_-_82477136 | 1.08 |
ENSRNOT00000026753
ENSRNOT00000089555 |
Resp18
|
regulated endocrine-specific protein 18 |
| chr11_+_45339366 | 0.97 |
ENSRNOT00000002241
|
Tmem30c
|
transmembrane protein 30C |
| chr8_+_20132900 | 0.89 |
ENSRNOT00000045980
|
AC099137.1
|
|
| chr9_-_8899144 | 0.82 |
ENSRNOT00000051043
|
AABR07066440.1
|
|
| chr10_-_87136026 | 0.78 |
ENSRNOT00000014230
ENSRNOT00000083233 |
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr11_-_87403367 | 0.77 |
ENSRNOT00000057751
|
Thap7
|
THAP domain containing 7 |
| chr4_+_31333970 | 0.74 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr13_-_70299733 | 0.72 |
ENSRNOT00000044235
|
Smg7
|
SMG7 nonsense mediated mRNA decay factor |
| chr1_-_54036068 | 0.69 |
ENSRNOT00000075019
|
RGD1560718
|
similar to putative protein kinase |
| chr8_+_41336340 | 0.61 |
ENSRNOT00000072049
|
Olr1225
|
olfactory receptor 1225 |
| chr9_+_66888393 | 0.60 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr5_+_36566783 | 0.58 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr4_-_66624912 | 0.53 |
ENSRNOT00000064891
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr10_+_63635219 | 0.52 |
ENSRNOT00000081813
ENSRNOT00000005016 |
Prpf8
|
pre-mRNA processing factor 8 |
| chr9_+_98438439 | 0.43 |
ENSRNOT00000027220
|
Scly
|
selenocysteine lyase |
| chr12_-_37910439 | 0.41 |
ENSRNOT00000001433
|
Ogfod2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr20_-_46044738 | 0.32 |
ENSRNOT00000000343
|
Fig4
|
FIG4 phosphoinositide 5-phosphatase |
| chr15_+_28695912 | 0.32 |
ENSRNOT00000081830
|
Tox4
|
TOX high mobility group box family member 4 |
| chr3_+_114900343 | 0.31 |
ENSRNOT00000068129
|
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr18_-_36579403 | 0.19 |
ENSRNOT00000025284
|
Lars
|
leucyl-tRNA synthetase |
| chr5_+_132005738 | 0.17 |
ENSRNOT00000041421
|
Skint4
|
selection and upkeep of intraepithelial T cells 4 |
| chr1_-_140835151 | 0.16 |
ENSRNOT00000032174
|
Hapln3
|
hyaluronan and proteoglycan link protein 3 |
| chr1_-_172552099 | 0.09 |
ENSRNOT00000090373
|
Olr257
|
olfactory receptor 257 |
| chr2_+_198721724 | 0.01 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Barhl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 16.9 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 3.1 | 9.2 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 2.4 | 9.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 2.2 | 6.7 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 2.1 | 8.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 2.1 | 14.4 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 1.9 | 5.7 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 1.8 | 5.4 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 1.4 | 4.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 1.3 | 7.6 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 1.2 | 6.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 1.1 | 10.8 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 1.0 | 3.1 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.9 | 7.8 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.8 | 3.2 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.7 | 8.9 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.6 | 2.4 | GO:0009597 | detection of virus(GO:0009597) |
| 0.5 | 1.6 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.5 | 5.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.4 | 2.8 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.3 | 2.1 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.3 | 6.3 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.3 | 1.8 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.3 | 2.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.3 | 2.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 1.8 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.2 | 5.0 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.2 | 3.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 3.4 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.1 | 1.7 | GO:1903140 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.1 | 1.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.5 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.4 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.6 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 1.4 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 1.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.7 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 3.2 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 2.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.9 | 9.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.8 | 3.1 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.8 | 14.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 3.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 4.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 2.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.2 | 2.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.2 | 9.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 1.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 8.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 5.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 4.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 8.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 16.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 6.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 8.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 2.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 6.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 7.8 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 4.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.2 | GO:0005903 | brush border(GO:0005903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.7 | 6.7 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 1.4 | 4.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 1.1 | 5.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.1 | 14.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 1.0 | 3.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 1.0 | 9.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.6 | 6.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.6 | 4.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.5 | 2.1 | GO:0001156 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.5 | 2.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.5 | 5.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.4 | 1.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.3 | 1.7 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 1.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 3.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.3 | 1.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.3 | 8.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.3 | 2.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 1.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 16.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 3.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 7.8 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 3.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 10.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 6.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 1.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 8.3 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 17.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 7.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.8 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 2.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.7 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 1.6 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 10.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 16.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 13.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 9.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 5.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 9.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.4 | 9.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 6.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.3 | 14.4 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.3 | 9.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 3.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 1.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 4.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 5.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 2.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 8.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 6.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |