Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Atoh1_Bhlhe23
Z-value: 0.55
Transcription factors associated with Atoh1_Bhlhe23
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
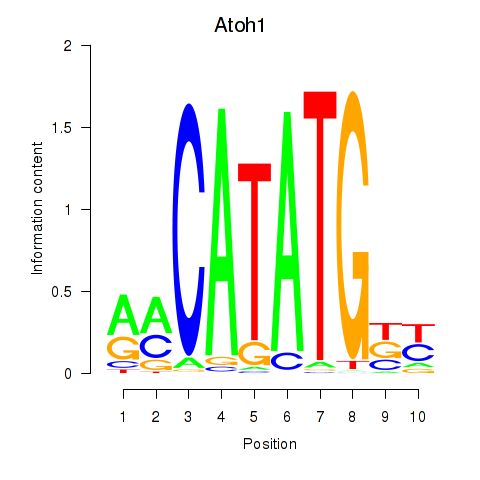
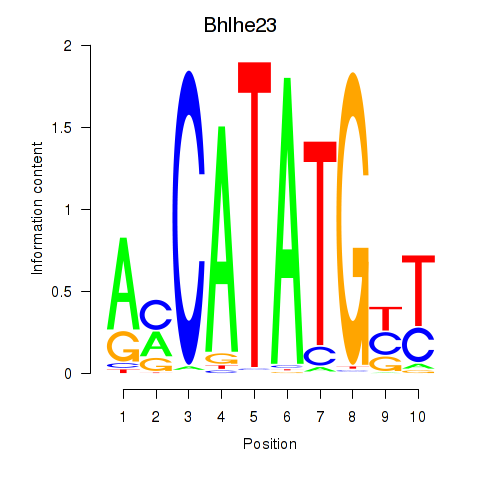
Atoh1
|
ENSRNOG00000006383 | atonal bHLH transcription factor 1 |
|
Bhlhe23
|
ENSRNOG00000010312 | basic helix-loop-helix family, member e23 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atoh1 | rn6_v1_chr4_+_95498003_95498003 | 0.15 | 6.3e-03 | Click! |
| Bhlhe23 | rn6_v1_chr3_-_176282211_176282211 | 0.00 | 9.8e-01 | Click! |
Activity profile of Atoh1_Bhlhe23 motif
Sorted Z-values of Atoh1_Bhlhe23 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_40038336 | 30.93 |
ENSRNOT00000048804
|
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr8_-_63750531 | 22.87 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr8_+_69971778 | 21.28 |
ENSRNOT00000058007
ENSRNOT00000037941 ENSRNOT00000050649 |
Megf11
|
multiple EGF-like-domains 11 |
| chr5_+_104362971 | 19.35 |
ENSRNOT00000058520
|
Adamtsl1
|
ADAMTS-like 1 |
| chr5_+_58636083 | 19.00 |
ENSRNOT00000067281
|
Unc13b
|
unc-13 homolog B |
| chr11_-_88972176 | 18.18 |
ENSRNOT00000002498
|
Pkp2
|
plakophilin 2 |
| chr16_+_2706428 | 16.59 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chr3_-_38090526 | 16.47 |
ENSRNOT00000059430
|
Cacnb4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr10_-_63952726 | 12.80 |
ENSRNOT00000090461
|
Doc2b
|
double C2 domain beta |
| chr16_-_32753278 | 11.80 |
ENSRNOT00000015759
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr6_+_60566196 | 11.59 |
ENSRNOT00000006709
ENSRNOT00000075193 |
Dock4
|
dedicator of cytokinesis 4 |
| chr11_+_72705129 | 11.25 |
ENSRNOT00000073330
|
Apod
|
apolipoprotein D |
| chr18_-_58423196 | 10.93 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr9_+_51298426 | 10.66 |
ENSRNOT00000065212
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr5_-_12563429 | 10.04 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr17_+_69468427 | 9.77 |
ENSRNOT00000058413
|
RGD1564865
|
similar to 20-alpha-hydroxysteroid dehydrogenase |
| chr4_-_31730386 | 8.32 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chr5_+_98469047 | 7.43 |
ENSRNOT00000041374
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr3_-_66417741 | 7.39 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr15_+_43905099 | 7.24 |
ENSRNOT00000016568
|
Ebf2
|
early B-cell factor 2 |
| chr2_-_100372252 | 7.16 |
ENSRNOT00000011890
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr4_+_122365093 | 6.76 |
ENSRNOT00000024011
|
Klf15
|
Kruppel-like factor 15 |
| chr19_+_20147037 | 6.56 |
ENSRNOT00000020028
|
Zfp423
|
zinc finger protein 423 |
| chr7_-_132343169 | 6.53 |
ENSRNOT00000021064
|
Abcd2
|
ATP binding cassette subfamily D member 2 |
| chr6_+_34030620 | 6.26 |
ENSRNOT00000091651
|
Laptm4a
|
lysosomal protein transmembrane 4 alpha |
| chr11_+_9642365 | 6.15 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr4_-_82194927 | 5.75 |
ENSRNOT00000072302
|
LOC103692128
|
homeobox protein Hox-A9 |
| chr11_-_87449940 | 5.72 |
ENSRNOT00000002560
|
Slc7a4
|
solute carrier family 7, member 4 |
| chr17_+_87274944 | 5.71 |
ENSRNOT00000073718
|
Etl4
|
enhancer trap locus 4 |
| chr4_-_173640684 | 5.65 |
ENSRNOT00000010728
|
Rergl
|
RERG like |
| chr2_-_184993341 | 5.62 |
ENSRNOT00000071580
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr5_-_12526962 | 5.56 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr7_-_52404774 | 5.55 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr3_+_37545238 | 5.30 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr3_+_45683993 | 4.82 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr6_+_107603580 | 4.67 |
ENSRNOT00000036430
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr2_+_207930796 | 4.63 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr7_-_117978787 | 4.43 |
ENSRNOT00000074262
|
Zfp647
|
zinc finger protein 647 |
| chr1_+_145715969 | 4.41 |
ENSRNOT00000037996
|
Tmc3
|
transmembrane channel-like 3 |
| chr7_-_12429897 | 4.41 |
ENSRNOT00000020670
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr13_+_82355886 | 4.28 |
ENSRNOT00000076757
|
Sele
|
selectin E |
| chr10_-_47724499 | 4.13 |
ENSRNOT00000085011
|
Rnf112
|
ring finger protein 112 |
| chr3_+_128756799 | 3.95 |
ENSRNOT00000049855
ENSRNOT00000042853 |
Plcb4
|
phospholipase C, beta 4 |
| chr5_-_79874671 | 3.94 |
ENSRNOT00000084563
|
Tnc
|
tenascin C |
| chrX_+_24891156 | 3.93 |
ENSRNOT00000071173
|
Wwc3
|
WWC family member 3 |
| chr14_+_22107416 | 3.74 |
ENSRNOT00000060179
ENSRNOT00000080114 |
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr1_+_84070983 | 3.70 |
ENSRNOT00000090478
|
Numbl
|
NUMB-like, endocytic adaptor protein |
| chr16_+_35573058 | 3.69 |
ENSRNOT00000059580
|
Galntl6
|
polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr10_+_69737328 | 3.66 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr16_+_23447366 | 3.64 |
ENSRNOT00000068629
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr7_+_13378338 | 3.63 |
ENSRNOT00000042747
|
Olr1073
|
olfactory receptor 1073 |
| chr14_-_3300200 | 3.37 |
ENSRNOT00000037931
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr3_+_20993205 | 3.26 |
ENSRNOT00000010465
|
Olr419
|
olfactory receptor 419 |
| chr10_+_59725398 | 3.05 |
ENSRNOT00000026156
|
P2rx5
|
purinergic receptor P2X 5 |
| chr10_+_84182118 | 3.01 |
ENSRNOT00000011027
|
Hoxb3
|
homeo box B3 |
| chr7_-_120839577 | 2.92 |
ENSRNOT00000018526
|
Dmc1
|
DNA meiotic recombinase 1 |
| chr1_-_72461547 | 2.77 |
ENSRNOT00000022480
|
Ssc5d
|
scavenger receptor cysteine rich family member with 5 domains |
| chr10_+_36517169 | 2.70 |
ENSRNOT00000040534
|
Olr1408
|
olfactory receptor 1408 |
| chr13_-_88642011 | 2.70 |
ENSRNOT00000067037
|
LOC100361087
|
hypothetical LOC100361087 |
| chr10_+_17542374 | 2.63 |
ENSRNOT00000064079
|
Fbxw11
|
F-box and WD repeat domain containing 11 |
| chr3_+_145764932 | 2.49 |
ENSRNOT00000075257
|
AABR07054264.1
|
|
| chr10_-_12674077 | 2.48 |
ENSRNOT00000041353
|
Olr1376
|
olfactory receptor 1376 |
| chr11_+_43394618 | 2.46 |
ENSRNOT00000051112
|
Olr1543
|
olfactory receptor 1543 |
| chr1_+_199351628 | 2.39 |
ENSRNOT00000078578
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr8_-_43443531 | 2.38 |
ENSRNOT00000007901
|
LOC103693060
|
olfactory receptor 148-like |
| chr2_+_40554146 | 2.31 |
ENSRNOT00000015138
|
Pde4d
|
phosphodiesterase 4D |
| chr15_-_12889102 | 2.23 |
ENSRNOT00000061214
ENSRNOT00000084687 |
RGD1306063
|
similar to HT021 |
| chr7_-_114848414 | 2.17 |
ENSRNOT00000032620
|
Slc45a4
|
solute carrier family 45, member 4 |
| chr20_+_3830164 | 2.07 |
ENSRNOT00000045533
ENSRNOT00000084117 |
Col11a2
|
collagen type XI alpha 2 chain |
| chr2_-_149444548 | 2.01 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr19_-_38321528 | 1.98 |
ENSRNOT00000031977
|
Smpd3
|
sphingomyelin phosphodiesterase 3 |
| chr15_-_100348760 | 1.92 |
ENSRNOT00000085607
|
AABR07019341.1
|
|
| chr3_+_172155496 | 1.83 |
ENSRNOT00000066279
|
Stx16
|
syntaxin 16 |
| chr2_+_115290243 | 1.82 |
ENSRNOT00000074802
|
LOC100910153
|
olfactory receptor 149-like |
| chr6_-_76552559 | 1.80 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chrX_+_156319687 | 1.80 |
ENSRNOT00000091147
ENSRNOT00000079378 |
Fam3a
|
family with sequence similarity 3, member A |
| chr4_-_170783725 | 1.75 |
ENSRNOT00000072528
|
Wbp11
|
WW domain binding protein 11 |
| chr1_+_168844875 | 1.70 |
ENSRNOT00000051398
|
AC107531.1
|
|
| chr8_+_41302631 | 1.64 |
ENSRNOT00000076963
|
LOC100910822
|
olfactory receptor 143-like |
| chr11_+_43309759 | 1.61 |
ENSRNOT00000040089
|
Olr1539
|
olfactory receptor 1539 |
| chr19_-_22632071 | 1.59 |
ENSRNOT00000077275
|
Gpt2
|
glutamic--pyruvic transaminase 2 |
| chr3_+_73307900 | 1.55 |
ENSRNOT00000051690
|
Olr469
|
olfactory receptor 469 |
| chr9_+_16302578 | 1.52 |
ENSRNOT00000021762
|
Gltscr1l
|
GLTSCR1-like |
| chr3_+_102947730 | 1.51 |
ENSRNOT00000071260
|
Olr773
|
olfactory receptor 773 |
| chr8_+_42663633 | 1.50 |
ENSRNOT00000048537
|
Olr1266
|
olfactory receptor 1266 |
| chrX_-_83864150 | 1.43 |
ENSRNOT00000073734
|
Obp1f
|
odorant binding protein I f |
| chr12_-_38504774 | 1.39 |
ENSRNOT00000011286
|
B3gnt4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chrX_+_122938009 | 1.36 |
ENSRNOT00000089266
ENSRNOT00000017550 |
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr4_+_7257752 | 1.36 |
ENSRNOT00000088357
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr1_+_230450529 | 1.36 |
ENSRNOT00000045383
|
Olr371
|
olfactory receptor 371 |
| chr1_-_205630073 | 1.35 |
ENSRNOT00000037064
|
Tex36
|
testis expressed 36 |
| chr1_+_150225373 | 1.30 |
ENSRNOT00000051266
|
Olr30
|
olfactory receptor 30 |
| chrX_+_1787266 | 1.25 |
ENSRNOT00000011183
|
Ndufb11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr15_+_27400717 | 1.21 |
ENSRNOT00000047008
|
Olr1620
|
olfactory receptor 1620 |
| chr1_-_238381871 | 1.19 |
ENSRNOT00000077601
|
Tmc1
|
transmembrane channel-like 1 |
| chr3_+_102579446 | 1.16 |
ENSRNOT00000047138
|
Olr754
|
olfactory receptor 754 |
| chr20_-_5754773 | 1.14 |
ENSRNOT00000035977
|
Ip6k3
|
inositol hexakisphosphate kinase 3 |
| chr3_+_72990358 | 1.13 |
ENSRNOT00000091920
|
Olr444
|
olfactory receptor 444 |
| chr7_-_118518193 | 1.12 |
ENSRNOT00000075636
|
LOC102555083
|
zinc finger protein 250-like |
| chr3_-_39596718 | 1.11 |
ENSRNOT00000006784
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr5_-_59266293 | 1.04 |
ENSRNOT00000050490
|
Olr836
|
olfactory receptor 836 |
| chr10_-_12818137 | 1.01 |
ENSRNOT00000060972
|
Olr1382
|
olfactory receptor 1382 |
| chr13_+_106751625 | 0.97 |
ENSRNOT00000004992
|
Ush2a
|
usherin |
| chr8_+_43382176 | 0.94 |
ENSRNOT00000047759
|
Olr1311
|
olfactory receptor 1311 |
| chr13_+_56096834 | 0.90 |
ENSRNOT00000035129
|
Dennd1b
|
DENN domain containing 1B |
| chr17_+_30556884 | 0.90 |
ENSRNOT00000080929
ENSRNOT00000022022 |
Eci2
|
enoyl-CoA delta isomerase 2 |
| chr2_+_196594303 | 0.89 |
ENSRNOT00000064442
ENSRNOT00000044738 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr18_+_80939875 | 0.88 |
ENSRNOT00000021729
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr3_-_151724654 | 0.88 |
ENSRNOT00000026964
|
Rbm39
|
RNA binding motif protein 39 |
| chr10_+_56260514 | 0.88 |
ENSRNOT00000016177
|
Sox15
|
SRY box 15 |
| chr1_+_230030914 | 0.87 |
ENSRNOT00000075730
|
Olr354
|
olfactory receptor 354 |
| chr1_-_228263198 | 0.87 |
ENSRNOT00000028572
|
Olr1874
|
olfactory receptor 1874 |
| chr3_+_15895191 | 0.86 |
ENSRNOT00000047879
|
Olr400
|
olfactory receptor 400 |
| chr5_+_137564607 | 0.85 |
ENSRNOT00000075742
|
LOC100912529
|
olfactory receptor 2B6-like |
| chr1_+_17602281 | 0.84 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr3_-_173839433 | 0.83 |
ENSRNOT00000075547
|
AC127963.1
|
|
| chr3_+_78876609 | 0.82 |
ENSRNOT00000049936
|
Olr726
|
olfactory receptor 726 |
| chr8_-_18062971 | 0.80 |
ENSRNOT00000045739
|
Olr1139
|
olfactory receptor 1139 |
| chr1_+_164425475 | 0.80 |
ENSRNOT00000023386
|
Klhl35
|
kelch-like family member 35 |
| chr11_+_43161181 | 0.79 |
ENSRNOT00000071470
|
Olr1530
|
olfactory receptor 1530 |
| chr8_+_43355060 | 0.78 |
ENSRNOT00000043206
|
Olr1309
|
olfactory receptor 1309 |
| chrX_+_74165187 | 0.71 |
ENSRNOT00000003973
|
Cdx4
|
caudal type homeo box 4 |
| chr7_-_13588411 | 0.70 |
ENSRNOT00000088783
|
Olr1083
|
olfactory receptor 1083 |
| chr3_-_152259156 | 0.66 |
ENSRNOT00000065729
|
LOC100910882
|
RNA-binding protein 39-like |
| chrX_-_76708878 | 0.65 |
ENSRNOT00000045534
|
Atrx
|
ATRX, chromatin remodeler |
| chr19_+_683484 | 0.64 |
ENSRNOT00000090332
|
Terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr3_+_55910177 | 0.63 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr11_+_43143882 | 0.60 |
ENSRNOT00000041445
|
Olr1528
|
olfactory receptor 1528 |
| chr17_-_57816256 | 0.59 |
ENSRNOT00000051570
|
LOC103694120
|
isopentenyl-diphosphate delta-isomerase 2-like |
| chr11_+_43476319 | 0.59 |
ENSRNOT00000044928
|
Olr1548
|
olfactory receptor 1548 |
| chr14_-_12387102 | 0.58 |
ENSRNOT00000038872
|
Bmp3
|
bone morphogenetic protein 3 |
| chr3_-_23353425 | 0.57 |
ENSRNOT00000019703
|
Golga1
|
golgin A1 |
| chr18_+_40883224 | 0.53 |
ENSRNOT00000039733
|
Lvrn
|
laeverin |
| chr2_+_196334626 | 0.51 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr10_+_60043848 | 0.49 |
ENSRNOT00000080593
|
AABR07029918.1
|
|
| chr16_+_55152748 | 0.48 |
ENSRNOT00000000121
|
Fgf20
|
fibroblast growth factor 20 |
| chr3_+_20965751 | 0.44 |
ENSRNOT00000010450
|
Olr417
|
olfactory receptor 417 |
| chr10_-_16045835 | 0.42 |
ENSRNOT00000064832
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr10_-_62366044 | 0.41 |
ENSRNOT00000045673
|
Olr1474
|
olfactory receptor 1474 |
| chr10_-_12644292 | 0.40 |
ENSRNOT00000041612
|
Olr1375
|
olfactory receptor 1375 |
| chr10_-_12370656 | 0.39 |
ENSRNOT00000060995
|
Olr1362
|
olfactory receptor 1362 |
| chr7_-_13531526 | 0.37 |
ENSRNOT00000048054
|
Olr1081
|
olfactory receptor 1081 |
| chr1_+_168830222 | 0.36 |
ENSRNOT00000050333
|
Olr122
|
olfactory receptor 122 |
| chr16_-_75028977 | 0.36 |
ENSRNOT00000058071
|
Defb13
|
defensin beta 13 |
| chr8_+_5790034 | 0.35 |
ENSRNOT00000061887
|
Mmp27
|
matrix metallopeptidase 27 |
| chr18_+_70192493 | 0.35 |
ENSRNOT00000020472
|
Cxxc1
|
CXXC finger protein 1 |
| chr7_+_125576327 | 0.33 |
ENSRNOT00000016679
|
Prr5
|
proline rich 5 |
| chr10_-_60772313 | 0.32 |
ENSRNOT00000050847
|
Olr1504
|
olfactory receptor 1504 |
| chr10_-_110308514 | 0.32 |
ENSRNOT00000054927
|
Tex19.2
|
testis expressed gene 19.2 |
| chr3_+_65816569 | 0.32 |
ENSRNOT00000079672
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr8_-_43082948 | 0.31 |
ENSRNOT00000072050
|
LOC100910667
|
olfactory receptor 8G5-like |
| chr8_+_41252456 | 0.28 |
ENSRNOT00000071683
|
Olr1220
|
olfactory receptor 1220 |
| chr10_-_12777245 | 0.25 |
ENSRNOT00000041708
|
Olr1380
|
olfactory receptor 1380 |
| chr3_-_66335869 | 0.21 |
ENSRNOT00000079781
ENSRNOT00000043238 ENSRNOT00000073412 ENSRNOT00000057878 ENSRNOT00000079212 |
Cerkl
|
ceramide kinase-like |
| chr3_-_73854363 | 0.20 |
ENSRNOT00000051169
|
Olr496
|
olfactory receptor 496 |
| chr3_-_74575010 | 0.19 |
ENSRNOT00000045231
|
Olr533
|
olfactory receptor 533 |
| chr11_+_43194348 | 0.18 |
ENSRNOT00000075303
|
Olr1532
|
olfactory receptor 1532 |
| chr1_+_213258684 | 0.18 |
ENSRNOT00000091678
|
LOC103690271
|
olfactory receptor 13A1-like |
| chr3_+_20878701 | 0.17 |
ENSRNOT00000046877
|
Olr413
|
olfactory receptor 413 |
| chr3_+_21016266 | 0.17 |
ENSRNOT00000010473
|
Olr420
|
olfactory receptor 420 |
| chr11_-_72086743 | 0.17 |
ENSRNOT00000072173
|
Pigz
|
phosphatidylinositol glycan anchor biosynthesis, class Z |
| chr1_+_230377079 | 0.13 |
ENSRNOT00000042937
|
Olr367
|
olfactory receptor 367 |
| chr7_-_13553963 | 0.10 |
ENSRNOT00000079450
|
Olr1082
|
olfactory receptor 1082 |
| chrX_-_76924362 | 0.09 |
ENSRNOT00000078997
|
Atrx
|
ATRX, chromatin remodeler |
| chr1_-_59903339 | 0.08 |
ENSRNOT00000039037
|
Fpr-rs3
|
formyl peptide receptor, related sequence 3 |
| chr2_-_210088949 | 0.07 |
ENSRNOT00000070994
|
Rbm15
|
RNA binding motif protein 15 |
| chr10_+_60728160 | 0.06 |
ENSRNOT00000041085
|
Olr1501
|
olfactory receptor 1501 |
| chr12_+_19561347 | 0.05 |
ENSRNOT00000060025
|
RGD1562319
|
similar to family with sequence similarity 55, member C |
| chr3_-_76085971 | 0.04 |
ENSRNOT00000007679
|
Olr601
|
olfactory receptor 601 |
| chr4_+_1482486 | 0.04 |
ENSRNOT00000035054
|
Olr1236
|
olfactory receptor 1236 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atoh1_Bhlhe23
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 3.8 | 11.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 3.6 | 18.2 | GO:0034334 | desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) |
| 2.7 | 19.0 | GO:0099525 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 2.1 | 8.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.1 | 6.2 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 1.5 | 22.9 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 1.5 | 7.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 1.3 | 16.5 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.2 | 3.7 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 1.1 | 12.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 1.0 | 3.9 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.8 | 6.5 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) very long-chain fatty acid catabolic process(GO:0042760) |
| 0.8 | 21.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.8 | 2.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.8 | 3.0 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.7 | 2.2 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.7 | 2.8 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.7 | 2.0 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.7 | 3.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.6 | 7.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.5 | 30.9 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.5 | 3.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 2.9 | GO:0042148 | strand invasion(GO:0042148) |
| 0.4 | 4.6 | GO:0098915 | potassium ion export across plasma membrane(GO:0097623) membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.4 | 3.9 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.3 | 1.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 2.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 2.0 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.3 | 6.8 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.3 | 1.6 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.2 | 0.7 | GO:0035128 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.2 | 1.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 4.8 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.2 | 10.7 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.2 | 2.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 6.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 1.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 3.9 | GO:0007602 | phototransduction(GO:0007602) |
| 0.2 | 4.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 3.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 4.3 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.1 | 2.6 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 1.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 10.9 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 4.1 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 1.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 5.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.9 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 0.9 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.6 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.6 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 2.0 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.6 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 1.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 5.7 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.9 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 1.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 7.0 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.4 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.6 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 1.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.5 | GO:0060043 | regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 0.7 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 20.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 19.0 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.8 | 17.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.7 | 2.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.6 | 4.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.5 | 2.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.4 | 18.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 2.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.3 | 13.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 14.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 0.7 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.2 | 4.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 6.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 3.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 3.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 6.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 4.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 32.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 8.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 41.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 2.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 4.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.9 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 2.9 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 11.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 30.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 2.1 | 19.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 2.1 | 22.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 2.1 | 16.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.7 | 8.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.4 | 18.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 1.3 | 12.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.9 | 4.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.9 | 16.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.7 | 2.2 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.6 | 3.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 3.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.5 | 1.6 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.5 | 6.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 2.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 1.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 3.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 4.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 0.9 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.3 | 2.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 2.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 11.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 0.9 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.2 | 0.9 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) |
| 0.2 | 4.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 2.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 10.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 5.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 12.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 3.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.2 | 7.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.6 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 3.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 7.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 5.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 3.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 3.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 33.7 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 11.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 3.1 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 16.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 3.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 6.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 11.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 2.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 19.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 19.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.7 | 36.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.6 | 22.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.5 | 14.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 16.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 6.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 3.9 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.3 | 6.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 4.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 8.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 2.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 2.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 17.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 2.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 11.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 4.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 2.6 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |