Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
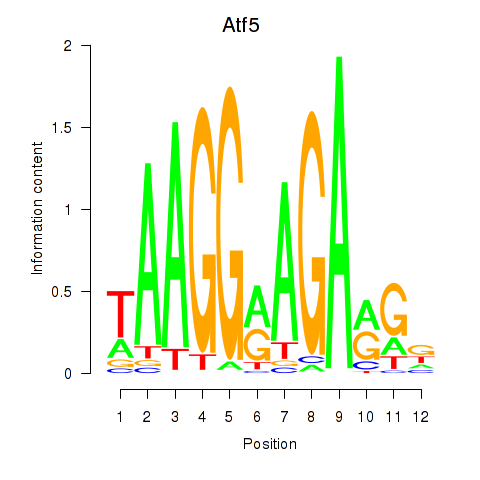
Results for Atf5
Z-value: 0.56
Transcription factors associated with Atf5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf5
|
ENSRNOG00000020060 | activating transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf5 | rn6_v1_chr1_-_100810522_100810522 | 0.02 | 6.7e-01 | Click! |
Activity profile of Atf5 motif
Sorted Z-values of Atf5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_39476384 | 21.37 |
ENSRNOT00000092968
|
Gpm6a
|
glycoprotein m6a |
| chr18_+_35574002 | 20.67 |
ENSRNOT00000093442
ENSRNOT00000070817 ENSRNOT00000093356 |
Myot
|
myotilin |
| chrX_-_29648359 | 20.36 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr9_+_37727942 | 20.22 |
ENSRNOT00000016511
ENSRNOT00000074276 |
LOC100912306
|
myotilin-like |
| chr16_-_39476025 | 19.92 |
ENSRNOT00000014312
|
Gpm6a
|
glycoprotein m6a |
| chr10_-_8498422 | 16.93 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr10_+_90348299 | 13.37 |
ENSRNOT00000089616
|
Rundc3a
|
RUN domain containing 3A |
| chr1_+_106998623 | 12.40 |
ENSRNOT00000022383
|
Slc17a6
|
solute carrier family 17 member 6 |
| chr9_-_60672246 | 10.93 |
ENSRNOT00000017761
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr18_+_13386133 | 10.73 |
ENSRNOT00000020661
|
Asxl3
|
additional sex combs like 3, transcriptional regulator |
| chr6_+_27768943 | 10.72 |
ENSRNOT00000015820
|
Kif3c
|
kinesin family member 3C |
| chr7_-_118332577 | 10.63 |
ENSRNOT00000090370
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr1_+_192379543 | 10.09 |
ENSRNOT00000078705
|
Prkcb
|
protein kinase C, beta |
| chr9_-_28972835 | 9.58 |
ENSRNOT00000086967
ENSRNOT00000079684 |
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr1_-_266074181 | 9.30 |
ENSRNOT00000026378
|
Psd
|
pleckstrin and Sec7 domain containing |
| chr10_+_86657285 | 8.37 |
ENSRNOT00000087346
|
Thra
|
thyroid hormone receptor alpha |
| chr20_-_5020150 | 7.95 |
ENSRNOT00000001146
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr16_-_7007287 | 7.28 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr7_+_79638046 | 7.22 |
ENSRNOT00000029419
|
Zfpm2
|
zinc finger protein, multitype 2 |
| chr1_+_245237736 | 6.91 |
ENSRNOT00000035814
|
Vldlr
|
very low density lipoprotein receptor |
| chr14_-_87701884 | 6.89 |
ENSRNOT00000079338
|
Mospd1
|
motile sperm domain containing 1 |
| chr1_+_228684136 | 6.65 |
ENSRNOT00000028608
|
Olr337
|
olfactory receptor 337 |
| chr2_-_220535751 | 6.51 |
ENSRNOT00000089082
|
Palmd
|
palmdelphin |
| chr12_+_41486076 | 6.47 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr13_-_76049363 | 6.41 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr1_+_199495298 | 6.35 |
ENSRNOT00000086003
ENSRNOT00000026748 |
Itgad
|
integrin subunit alpha D |
| chr15_+_52557607 | 6.13 |
ENSRNOT00000019222
|
Gfra2
|
GDNF family receptor alpha 2 |
| chr3_+_116899878 | 6.05 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr9_-_71788281 | 6.03 |
ENSRNOT00000020078
|
Crygc
|
crystallin, gamma C |
| chr5_-_21345805 | 6.02 |
ENSRNOT00000081296
ENSRNOT00000007802 |
Car8
|
carbonic anhydrase 8 |
| chr11_-_31180616 | 5.46 |
ENSRNOT00000065535
ENSRNOT00000052033 ENSRNOT00000002812 |
Synj1
|
synaptojanin 1 |
| chr1_+_46873011 | 5.45 |
ENSRNOT00000068476
ENSRNOT00000067661 |
Synj2
|
synaptojanin 2 |
| chr12_+_25036605 | 5.41 |
ENSRNOT00000001996
ENSRNOT00000084427 |
Limk1
|
LIM domain kinase 1 |
| chr8_+_33239139 | 5.39 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr13_+_43818010 | 5.09 |
ENSRNOT00000005532
|
LOC100909664
|
centrosomal protein of 170 kDa-like |
| chr8_+_52751854 | 4.93 |
ENSRNOT00000072518
|
Nxpe1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr1_-_198233215 | 4.77 |
ENSRNOT00000087928
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr13_-_94859390 | 4.76 |
ENSRNOT00000005467
ENSRNOT00000079763 |
AABR07021840.1
|
|
| chr1_-_57815038 | 4.66 |
ENSRNOT00000075401
|
Rgmb
|
repulsive guidance molecule family member B |
| chr1_+_198284473 | 4.37 |
ENSRNOT00000027022
|
Doc2a
|
double C2 domain alpha |
| chr9_-_45206427 | 4.06 |
ENSRNOT00000042227
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr2_+_216863428 | 3.83 |
ENSRNOT00000068413
|
Col11a1
|
collagen type XI alpha 1 chain |
| chr16_+_27399467 | 3.82 |
ENSRNOT00000065642
|
Tll1
|
tolloid-like 1 |
| chr1_-_255815733 | 3.79 |
ENSRNOT00000047387
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr1_-_103380338 | 3.35 |
ENSRNOT00000019061
|
Mrgprx4
|
MAS related GPR family member X4 |
| chr5_-_138462864 | 2.38 |
ENSRNOT00000011325
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr7_+_80750725 | 2.18 |
ENSRNOT00000079962
|
Oxr1
|
oxidation resistance 1 |
| chrX_+_5549419 | 2.09 |
ENSRNOT00000004676
|
Fundc1
|
FUN14 domain containing 1 |
| chr1_+_72399157 | 1.92 |
ENSRNOT00000022349
|
Zfp579
|
zinc finger protein 579 |
| chr1_-_7480825 | 1.89 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr8_-_131899023 | 1.67 |
ENSRNOT00000034690
|
Zfp445
|
zinc finger protein 445 |
| chr1_+_221872220 | 1.59 |
ENSRNOT00000028651
|
Nrxn2
|
neurexin 2 |
| chr13_-_82758004 | 1.59 |
ENSRNOT00000003932
|
Atp1b1
|
ATPase Na+/K+ transporting subunit beta 1 |
| chr1_-_169845487 | 1.46 |
ENSRNOT00000039989
|
Olr179
|
olfactory receptor 179 |
| chr9_+_10044756 | 1.45 |
ENSRNOT00000071188
|
Pspn
|
persephin |
| chr3_+_75999481 | 1.44 |
ENSRNOT00000074642
|
Olr595
|
olfactory receptor 595 |
| chr9_-_16643182 | 1.42 |
ENSRNOT00000024266
|
LOC108348250
|
cullin-7 |
| chr1_+_229443102 | 1.39 |
ENSRNOT00000067320
|
Olr334
|
olfactory receptor 334 |
| chr5_+_133864798 | 1.32 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr3_-_79165698 | 1.31 |
ENSRNOT00000032617
|
Olr744
|
olfactory receptor 744 |
| chr10_-_12674077 | 1.23 |
ENSRNOT00000041353
|
Olr1376
|
olfactory receptor 1376 |
| chr10_-_12312692 | 1.22 |
ENSRNOT00000073059
|
Olr1355
|
olfactory receptor 1355 |
| chr3_+_15688992 | 1.18 |
ENSRNOT00000045037
|
Olr395
|
olfactory receptor 395 |
| chr19_+_9024451 | 1.09 |
ENSRNOT00000073744
|
LOC100910721
|
60S ribosomal protein L26-like |
| chr2_+_188141350 | 1.08 |
ENSRNOT00000078913
|
Gon4l
|
gon-4 like |
| chr13_-_76660043 | 1.06 |
ENSRNOT00000089332
|
Pappa2
|
pappalysin 2 |
| chr1_+_198682230 | 1.02 |
ENSRNOT00000023995
|
Znf48
|
zinc finger protein 48 |
| chr3_+_75768699 | 0.95 |
ENSRNOT00000071912
|
AABR07052798.1
|
|
| chr6_-_99204825 | 0.93 |
ENSRNOT00000043602
|
Esr2
|
estrogen receptor 2 |
| chr2_-_35192515 | 0.85 |
ENSRNOT00000086631
|
LOC100909937
|
olfactory receptor 149-like |
| chr1_-_89944752 | 0.84 |
ENSRNOT00000029680
|
Scgb2b24
|
secretoglobin, family 2B, member 24 |
| chr1_+_68670127 | 0.79 |
ENSRNOT00000043828
|
Vom1r63
|
vomeronasal 1 receptor 63 |
| chr6_-_99204590 | 0.65 |
ENSRNOT00000041872
ENSRNOT00000007109 |
Esr2
|
estrogen receptor 2 |
| chr3_-_80601410 | 0.64 |
ENSRNOT00000022841
|
Atg13
|
autophagy related 13 |
| chr7_-_121943121 | 0.55 |
ENSRNOT00000046460
|
Rpl26-ps1
|
ribosomal protein L26, pseudogene 1 |
| chr3_+_15895191 | 0.54 |
ENSRNOT00000047879
|
Olr400
|
olfactory receptor 400 |
| chr3_+_172510874 | 0.49 |
ENSRNOT00000073634
|
Nelfcd
|
negative elongation factor complex member C/D |
| chr1_-_31528083 | 0.39 |
ENSRNOT00000061041
|
Rpl26-ps2
|
ribosomal protein L26, pseudogene 2 |
| chr13_-_76660245 | 0.25 |
ENSRNOT00000039210
|
Pappa2
|
pappalysin 2 |
| chr4_+_60549197 | 0.24 |
ENSRNOT00000071249
ENSRNOT00000075621 |
Exoc4
|
exocyst complex component 4 |
| chr17_+_45801528 | 0.16 |
ENSRNOT00000089221
|
Olr1664
|
olfactory receptor 1664 |
| chr5_-_173209768 | 0.15 |
ENSRNOT00000045053
|
Atad3a
|
ATPase family, AAA domain containing 3A |
| chr3_-_73493621 | 0.14 |
ENSRNOT00000071782
|
Olr482
|
olfactory receptor 482 |
| chr9_+_99735011 | 0.13 |
ENSRNOT00000051413
|
Olr1347
|
olfactory receptor 1347 |
| chr1_+_200037749 | 0.10 |
ENSRNOT00000027630
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr3_+_15930518 | 0.06 |
ENSRNOT00000040288
|
Olr401
|
olfactory receptor 401 |
| chr10_+_12602388 | 0.01 |
ENSRNOT00000060989
|
Olr1373
|
olfactory receptor 1373 |
| chr3_+_16287056 | 0.01 |
ENSRNOT00000045825
|
Olr408
|
olfactory receptor 408 |
| chr1_+_149963547 | 0.01 |
ENSRNOT00000042245
|
Olr20
|
olfactory receptor 20 |
| chrM_+_2740 | 0.00 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 20.4 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 3.4 | 10.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 3.2 | 9.6 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 2.8 | 8.4 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 2.4 | 7.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 1.7 | 6.9 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 1.6 | 12.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 1.6 | 9.3 | GO:0006863 | purine nucleobase transport(GO:0006863) nucleobase transport(GO:0015851) |
| 1.5 | 10.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.5 | 13.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 1.1 | 5.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.0 | 6.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 1.0 | 41.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.9 | 3.8 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.7 | 6.4 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.7 | 16.9 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.5 | 1.6 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.4 | 1.3 | GO:0045799 | basophil differentiation(GO:0030221) positive regulation of chromatin assembly or disassembly(GO:0045799) regulation of mast cell differentiation(GO:0060375) |
| 0.4 | 3.8 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 1.6 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.4 | 5.4 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.3 | 4.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 1.6 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.2 | 10.9 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.2 | 7.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 2.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 5.4 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.2 | 6.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 6.0 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.2 | 6.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 3.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 10.7 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.1 | 4.1 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 4.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 6.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 4.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 4.7 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 5.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.6 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 2.1 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 1.4 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 10.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 1.3 | 3.8 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 1.2 | 9.6 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.1 | 5.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 1.0 | 46.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.5 | 12.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 6.9 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.3 | 5.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.2 | 1.4 | GO:1990393 | Cul7-RING ubiquitin ligase complex(GO:0031467) 3M complex(GO:1990393) |
| 0.2 | 3.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 25.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 10.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 10.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 4.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 16.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 15.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 2.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 2.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 3.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 20.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 5.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 8.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 5.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 2.3 | 6.9 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 1.9 | 13.4 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 1.8 | 5.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.8 | 5.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.8 | 12.4 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.8 | 9.3 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.8 | 8.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.7 | 4.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.5 | 6.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 6.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 20.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.4 | 1.6 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.3 | 3.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 41.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.3 | 6.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 4.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 1.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 10.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.6 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 27.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 11.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 9.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 4.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 7.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 6.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 10.2 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 3.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0034595 | phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 6.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 6.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 3.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 6.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 8.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 6.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 5.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 5.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 12.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 7.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.5 | 10.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 10.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 6.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 10.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.3 | 5.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 12.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 6.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 9.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 5.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 4.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 4.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 7.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |