Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
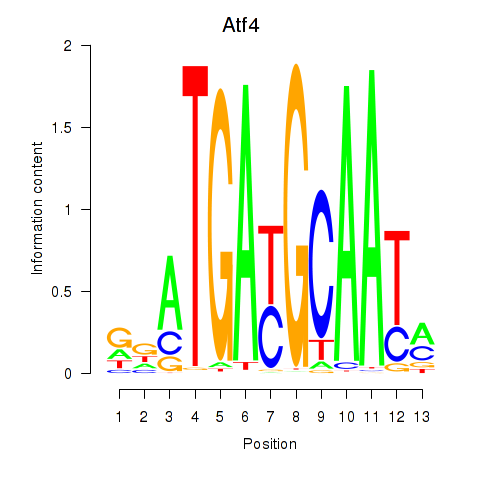
Results for Atf4
Z-value: 0.87
Transcription factors associated with Atf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf4
|
ENSRNOG00000017801 | activating transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf4 | rn6_v1_chr7_+_121480723_121480723 | -0.16 | 5.1e-03 | Click! |
Activity profile of Atf4 motif
Sorted Z-values of Atf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_70643169 | 44.36 |
ENSRNOT00000010106
|
Inhbe
|
inhibin beta E subunit |
| chr3_+_111160205 | 42.53 |
ENSRNOT00000019392
|
Chac1
|
ChaC glutathione-specific gamma-glutamylcyclotransferase 1 |
| chr1_-_101596822 | 31.00 |
ENSRNOT00000028490
|
Fgf21
|
fibroblast growth factor 21 |
| chr9_+_74124016 | 30.01 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr10_-_62254287 | 21.53 |
ENSRNOT00000004313
|
Serpinf1
|
serpin family F member 1 |
| chr5_-_137372993 | 19.54 |
ENSRNOT00000092823
|
Tmem125
|
transmembrane protein 125 |
| chr1_-_194769524 | 16.74 |
ENSRNOT00000025988
|
Nupr1
|
nuclear protein 1, transcriptional regulator |
| chr1_-_198045154 | 15.99 |
ENSRNOT00000072875
|
Nupr1l1
|
nuclear protein, transcriptional regulator, 1-like 1 |
| chr7_-_70829815 | 15.85 |
ENSRNOT00000011082
|
Shmt2
|
serine hydroxymethyltransferase 2 |
| chr13_+_103300932 | 13.61 |
ENSRNOT00000085214
ENSRNOT00000038264 |
Eprs
|
glutamyl-prolyl-tRNA synthetase |
| chr5_+_136669674 | 13.36 |
ENSRNOT00000048770
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr1_-_263269762 | 12.89 |
ENSRNOT00000022309
|
Got1
|
glutamic-oxaloacetic transaminase 1 |
| chr19_-_43215281 | 12.70 |
ENSRNOT00000025052
|
Aars
|
alanyl-tRNA synthetase |
| chr9_-_78969013 | 12.39 |
ENSRNOT00000019772
ENSRNOT00000057585 |
Fn1
|
fibronectin 1 |
| chr1_-_216801656 | 12.14 |
ENSRNOT00000050721
|
Cars
|
cysteinyl-tRNA synthetase |
| chr19_-_43215077 | 12.04 |
ENSRNOT00000082151
|
Aars
|
alanyl-tRNA synthetase |
| chr7_+_130296897 | 12.04 |
ENSRNOT00000044854
|
Adm2
|
adrenomedullin 2 |
| chr19_-_11057254 | 11.95 |
ENSRNOT00000025559
|
Herpud1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr11_-_29641551 | 11.84 |
ENSRNOT00000064103
|
Krtap7-1
|
keratin associated protein 7-1 |
| chr14_+_22072024 | 11.35 |
ENSRNOT00000002680
|
ste2
|
estrogen sulfotransferase |
| chr10_-_38782419 | 10.92 |
ENSRNOT00000073964
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr10_-_109821807 | 10.49 |
ENSRNOT00000054949
|
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr5_+_147375350 | 10.31 |
ENSRNOT00000010674
|
Yars
|
tyrosyl-tRNA synthetase |
| chr7_+_141054494 | 9.96 |
ENSRNOT00000088176
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr13_+_49933155 | 9.55 |
ENSRNOT00000037039
|
Ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr8_+_22856539 | 9.05 |
ENSRNOT00000015381
|
Angptl8
|
angiopoietin-like 8 |
| chr7_-_101140308 | 7.95 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr15_-_35113678 | 7.51 |
ENSRNOT00000059677
|
Ctsg
|
cathepsin G |
| chr14_+_89384512 | 6.62 |
ENSRNOT00000035612
|
Abca13
|
ATP binding cassette subfamily A member 13 |
| chr1_-_91296656 | 5.98 |
ENSRNOT00000028703
|
Cebpg
|
CCAAT/enhancer binding protein gamma |
| chr11_+_80319177 | 5.83 |
ENSRNOT00000036777
|
Rtp2
|
receptor (chemosensory) transporter protein 2 |
| chr4_+_85235172 | 5.63 |
ENSRNOT00000014780
|
Gars
|
glycyl-tRNA synthetase |
| chr20_+_5933303 | 5.54 |
ENSRNOT00000000617
|
Mapk14
|
mitogen activated protein kinase 14 |
| chr4_-_44136815 | 5.46 |
ENSRNOT00000086810
|
Tfec
|
transcription factor EC |
| chr19_-_11451278 | 5.30 |
ENSRNOT00000026118
|
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chrX_-_130794673 | 5.24 |
ENSRNOT00000080612
|
LOC691030
|
similar to RIKEN cDNA 1700001F22 |
| chr17_-_15402631 | 5.24 |
ENSRNOT00000019862
ENSRNOT00000093637 |
Iars
|
isoleucyl-tRNA synthetase |
| chr10_-_20622600 | 4.73 |
ENSRNOT00000020554
|
Fbll1
|
fibrillarin-like 1 |
| chr5_+_59452348 | 4.71 |
ENSRNOT00000019887
|
Ccin
|
calicin |
| chr4_-_157829241 | 4.47 |
ENSRNOT00000026184
|
Ltbr
|
lymphotoxin beta receptor |
| chr19_+_14508616 | 4.46 |
ENSRNOT00000019192
|
Hmox1
|
heme oxygenase 1 |
| chr2_-_189333015 | 4.38 |
ENSRNOT00000076424
ENSRNOT00000076952 |
Hax1
|
HCLS1 associated protein X-1 |
| chr2_-_189333322 | 4.35 |
ENSRNOT00000073599
ENSRNOT00000071253 ENSRNOT00000076325 |
Hax1
|
HCLS1 associated protein X-1 |
| chr8_-_13680870 | 4.33 |
ENSRNOT00000081616
|
Hephl1
|
hephaestin-like 1 |
| chr10_+_84119884 | 4.07 |
ENSRNOT00000009951
|
Hoxb9
|
homeo box B9 |
| chrX_-_144001727 | 3.95 |
ENSRNOT00000078404
|
RGD1560585
|
similar to RIKEN cDNA 1700001F22 |
| chr10_-_75120247 | 3.77 |
ENSRNOT00000011402
|
Lpo
|
lactoperoxidase |
| chr12_+_48677905 | 3.77 |
ENSRNOT00000083196
|
1700069L16Rik
|
RIKEN cDNA 1700069L16 gene |
| chr17_-_53087274 | 3.62 |
ENSRNOT00000020794
|
RGD1308147
|
similar to expressed sequence AW209491 |
| chr4_+_133540540 | 3.61 |
ENSRNOT00000073232
|
AABR07061556.1
|
|
| chr8_-_48674748 | 3.54 |
ENSRNOT00000014127
|
Hmbs
|
hydroxymethylbilane synthase |
| chr4_-_115015965 | 3.54 |
ENSRNOT00000014603
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr9_-_84383687 | 3.41 |
ENSRNOT00000019685
|
Farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr11_-_28842287 | 3.32 |
ENSRNOT00000061610
|
Krtap19-5
|
keratin associated protein 19-5 |
| chr1_+_102849889 | 3.23 |
ENSRNOT00000066791
|
Gtf2h1
|
general transcription factor IIH subunit 1 |
| chr19_+_49695579 | 3.13 |
ENSRNOT00000016963
|
Gan
|
gigaxonin |
| chr14_+_96300596 | 3.12 |
ENSRNOT00000071967
|
LOC302228
|
similar to Spindlin-like protein 2 (SPIN-2) |
| chr1_+_174655939 | 3.01 |
ENSRNOT00000014002
|
Ipo7
|
importin 7 |
| chr18_-_36579403 | 2.88 |
ENSRNOT00000025284
|
Lars
|
leucyl-tRNA synthetase |
| chr12_+_11240761 | 2.83 |
ENSRNOT00000001310
|
Pdap1
|
PDGFA associated protein 1 |
| chr15_+_30735921 | 2.73 |
ENSRNOT00000078887
|
AABR07017763.2
|
|
| chr18_-_60002529 | 2.66 |
ENSRNOT00000081014
ENSRNOT00000024264 ENSRNOT00000059162 |
Nars
|
asparaginyl-tRNA synthetase |
| chr4_+_163293724 | 2.66 |
ENSRNOT00000077356
|
Gabarapl1
|
GABA type A receptor associated protein like 1 |
| chr16_+_8207223 | 2.52 |
ENSRNOT00000026751
|
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr18_-_67224566 | 2.30 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr3_-_77911561 | 2.20 |
ENSRNOT00000084947
|
Olr679
|
olfactory receptor 679 |
| chr4_+_145440114 | 2.15 |
ENSRNOT00000013052
|
Creld1
|
cysteine-rich with EGF-like domains 1 |
| chr18_+_40596166 | 1.99 |
ENSRNOT00000087984
|
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr1_-_233145924 | 1.88 |
ENSRNOT00000018763
ENSRNOT00000082442 |
Psat1
|
phosphoserine aminotransferase 1 |
| chr4_-_68349273 | 1.82 |
ENSRNOT00000016251
|
Prss37
|
protease, serine, 37 |
| chr6_+_64808238 | 1.74 |
ENSRNOT00000093195
|
Nrcam
|
neuronal cell adhesion molecule |
| chr1_+_100473643 | 1.74 |
ENSRNOT00000026379
|
Josd2
|
Josephin domain containing 2 |
| chr16_-_70998575 | 1.66 |
ENSRNOT00000019935
|
Kcnu1
|
potassium calcium-activated channel subfamily U member 1 |
| chr16_-_8207464 | 1.45 |
ENSRNOT00000026742
|
Dph3
|
diphthamide biosynthesis 3 |
| chr7_+_123381077 | 1.38 |
ENSRNOT00000082603
ENSRNOT00000056041 |
Srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr7_+_18310624 | 1.16 |
ENSRNOT00000075258
|
Actl9
|
actin-like 9 |
| chr2_-_31753528 | 1.09 |
ENSRNOT00000075057
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr19_+_53629779 | 0.94 |
ENSRNOT00000051352
|
Map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr6_+_18821815 | 0.93 |
ENSRNOT00000062057
|
Mtm1
|
myotubularin 1 |
| chr15_-_33527031 | 0.86 |
ENSRNOT00000019979
|
Homez
|
homeobox and leucine zipper encoding |
| chr19_+_37668133 | 0.76 |
ENSRNOT00000044971
|
Pard6a
|
par-6 family cell polarity regulator alpha |
| chr7_-_63728988 | 0.53 |
ENSRNOT00000009645
|
Xpot
|
exportin for tRNA |
| chr10_+_57239993 | 0.46 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr7_-_117773134 | 0.22 |
ENSRNOT00000045135
|
Recql4
|
RecQ like helicase 4 |
| chr19_+_37668693 | 0.11 |
ENSRNOT00000084970
|
Pard6a
|
par-6 family cell polarity regulator alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 31.0 | GO:1904640 | response to methionine(GO:1904640) |
| 6.2 | 24.7 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 6.0 | 30.0 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 5.3 | 42.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 4.5 | 13.6 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 4.3 | 12.9 | GO:0006532 | fumarate metabolic process(GO:0006106) glycerol biosynthetic process(GO:0006114) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 4.1 | 12.4 | GO:2001201 | calcium-independent cell-matrix adhesion(GO:0007161) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 3.6 | 10.9 | GO:0021539 | subthalamus development(GO:0021539) |
| 3.5 | 17.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 3.5 | 10.5 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 3.3 | 10.0 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 2.5 | 7.5 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 2.4 | 21.5 | GO:0071279 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 2.4 | 12.0 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 1.9 | 5.6 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 1.6 | 4.7 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 1.5 | 4.5 | GO:0006788 | heme oxidation(GO:0006788) |
| 1.4 | 5.5 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 1.4 | 9.5 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 1.3 | 5.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 1.2 | 13.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 1.1 | 16.7 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 1.0 | 2.9 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.9 | 2.7 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.9 | 5.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.8 | 25.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.8 | 44.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.8 | 8.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.7 | 6.0 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.6 | 9.1 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 0.6 | 3.5 | GO:0018065 | protein-cofactor linkage(GO:0018065) cellular response to amine stimulus(GO:0071418) |
| 0.6 | 2.3 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.3 | 1.4 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 9.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 3.5 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.2 | 0.9 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.2 | 1.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 13.1 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.2 | 0.5 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 4.5 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 3.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.9 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 5.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.9 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 1.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 4.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 4.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 2.0 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.4 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 1.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 3.0 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.0 | GO:1990037 | Lewy body core(GO:1990037) |
| 3.2 | 15.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 2.7 | 21.5 | GO:0043203 | axon hillock(GO:0043203) |
| 2.1 | 26.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 1.5 | 12.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.1 | 3.4 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 1.0 | 10.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.7 | 9.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.6 | 4.7 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 30.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.5 | 1.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 1.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.4 | 3.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 2.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 12.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 42.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 8.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 7.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 12.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 4.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 13.4 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 10.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 5.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 54.2 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 37.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.9 | GO:0030175 | filopodium(GO:0030175) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.6 | 42.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 6.2 | 24.7 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 5.3 | 15.8 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 4.5 | 13.6 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 4.3 | 12.9 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 3.5 | 10.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 2.5 | 30.0 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 2.5 | 10.0 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.8 | 12.4 | GO:0045340 | mercury ion binding(GO:0045340) |
| 1.5 | 4.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 1.4 | 10.9 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.3 | 5.3 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 1.3 | 5.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 1.2 | 3.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.2 | 5.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 1.0 | 13.4 | GO:0015187 | sodium:amino acid symporter activity(GO:0005283) glycine transmembrane transporter activity(GO:0015187) |
| 1.0 | 31.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 1.0 | 2.9 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.9 | 34.1 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.9 | 3.5 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.8 | 44.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.7 | 8.7 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.7 | 4.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.7 | 5.5 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) NFAT protein binding(GO:0051525) |
| 0.5 | 2.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.4 | 1.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.4 | 4.7 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.3 | 2.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 6.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 4.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 2.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 1.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 0.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 21.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 3.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 9.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 21.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 3.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 3.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 1.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 12.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 7.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0000405 | bubble DNA binding(GO:0000405) four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 7.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 6.6 | GO:0016887 | ATPase activity(GO:0016887) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 19.0 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 72.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 5.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 7.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 21.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 5.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 44.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 2.9 | 77.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 1.4 | 25.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.7 | 12.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.6 | 13.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 8.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.5 | 7.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.4 | 7.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 7.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 10.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 30.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 4.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 3.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |