Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
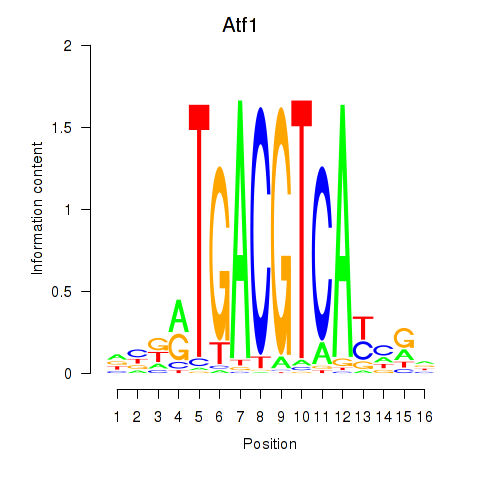
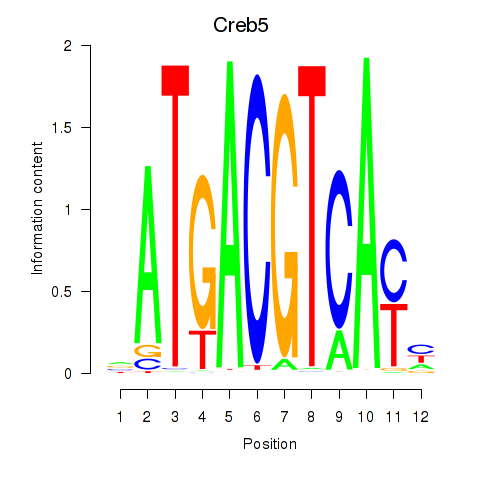
Results for Atf1_Creb5
Z-value: 0.87
Transcription factors associated with Atf1_Creb5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf1
|
ENSRNOG00000061088 | activating transcription factor 1 |
|
Creb5
|
ENSRNOG00000008622 | cAMP responsive element binding protein 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf1 | rn6_v1_chr7_+_141889444_141889444 | 0.31 | 2.3e-08 | Click! |
| Creb5 | rn6_v1_chr4_+_83713666_83713666 | -0.21 | 2.1e-04 | Click! |
Activity profile of Atf1_Creb5 motif
Sorted Z-values of Atf1_Creb5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_113358526 | 56.53 |
ENSRNOT00000065073
|
Txndc2
|
thioredoxin domain containing 2 |
| chr5_-_146446227 | 55.22 |
ENSRNOT00000044868
|
Hmgb4
|
high-mobility group box 4 |
| chr8_-_115263484 | 48.88 |
ENSRNOT00000039696
ENSRNOT00000081138 |
Iqcf3
|
IQ motif containing F3 |
| chr8_-_115274165 | 47.63 |
ENSRNOT00000056386
|
LOC102550160
|
IQ domain-containing protein F5-like |
| chr5_-_140024176 | 47.36 |
ENSRNOT00000016492
|
Tmco2
|
transmembrane and coiled-coil domains 2 |
| chr8_+_115213471 | 47.05 |
ENSRNOT00000017570
|
Iqcf5
|
IQ motif containing F5 |
| chr7_+_18310624 | 45.95 |
ENSRNOT00000075258
|
Actl9
|
actin-like 9 |
| chr1_-_89190128 | 41.74 |
ENSRNOT00000067813
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr6_-_86223052 | 41.21 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr5_+_59452348 | 40.96 |
ENSRNOT00000019887
|
Ccin
|
calicin |
| chr5_+_73496027 | 40.10 |
ENSRNOT00000022395
|
Actl7a
|
actin-like 7a |
| chr15_-_5417570 | 39.91 |
ENSRNOT00000061525
|
Spetex-2F
|
Spetex-2F protein |
| chr4_-_6449949 | 39.53 |
ENSRNOT00000062026
|
Galntl5
|
polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr16_-_82288022 | 38.38 |
ENSRNOT00000078609
|
Spaca7
|
sperm acrosome associated 7 |
| chrX_-_144001727 | 37.26 |
ENSRNOT00000078404
|
RGD1560585
|
similar to RIKEN cDNA 1700001F22 |
| chr15_+_28319136 | 36.53 |
ENSRNOT00000048723
|
Tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr1_-_198267093 | 35.72 |
ENSRNOT00000047477
|
RGD1563217
|
similar to RIKEN cDNA 4930451I11 |
| chr11_+_88424414 | 35.31 |
ENSRNOT00000022328
|
Spag6l
|
sperm associated antigen 6-like |
| chr1_-_24604400 | 34.13 |
ENSRNOT00000081175
|
1700020N01Rik
|
RIKEN cDNA 1700020N01 gene |
| chr8_+_115193146 | 33.05 |
ENSRNOT00000056391
|
Iqcf1
|
IQ motif containing F1 |
| chrX_-_132424746 | 32.84 |
ENSRNOT00000087819
|
LOC681067
|
similar to RIKEN cDNA 1700001F22 |
| chrX_-_139329975 | 32.46 |
ENSRNOT00000086405
|
LOC103694517
|
high mobility group protein B4-like |
| chrX_-_130794673 | 32.06 |
ENSRNOT00000080612
|
LOC691030
|
similar to RIKEN cDNA 1700001F22 |
| chr4_-_169036950 | 31.95 |
ENSRNOT00000011295
|
Gsg1
|
germ cell associated 1 |
| chr16_+_83161880 | 31.92 |
ENSRNOT00000080793
|
1700016D06Rik
|
RIKEN cDNA 1700016D06 gene |
| chr9_-_85243001 | 31.42 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr6_+_28515025 | 31.28 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chrX_+_137197396 | 29.25 |
ENSRNOT00000082227
|
RGD1566225
|
similar to RIKEN cDNA 1700001F22 |
| chr2_+_240396152 | 28.79 |
ENSRNOT00000034565
|
Cenpe
|
centromere protein E |
| chr6_-_47848026 | 28.76 |
ENSRNOT00000011048
ENSRNOT00000090017 |
Allc
|
allantoicase |
| chr6_+_122603269 | 28.64 |
ENSRNOT00000005297
|
Spata7
|
spermatogenesis associated 7 |
| chr3_+_170475831 | 27.98 |
ENSRNOT00000006949
|
Fam209a
|
family with sequence similarity 209, member A |
| chr6_+_126434226 | 27.81 |
ENSRNOT00000090857
|
Chga
|
chromogranin A |
| chrX_-_158261717 | 27.48 |
ENSRNOT00000086804
|
RGD1561558
|
similar to RIKEN cDNA 1700001F22 |
| chr9_-_63291350 | 27.02 |
ENSRNOT00000058831
|
Hsfy2
|
heat shock transcription factor, Y linked 2 |
| chr15_-_5467047 | 26.40 |
ENSRNOT00000042743
|
Spetex-2F
|
Spetex-2F protein |
| chr6_+_26780352 | 26.07 |
ENSRNOT00000009916
|
Prr30
|
proline rich 30 |
| chr3_+_35014538 | 25.70 |
ENSRNOT00000006341
|
Kif5c
|
kinesin family member 5C |
| chrX_+_114929029 | 25.55 |
ENSRNOT00000006459
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr1_-_169334093 | 25.19 |
ENSRNOT00000032587
|
Ubqln3
|
ubiquilin 3 |
| chr9_+_82053581 | 24.94 |
ENSRNOT00000086375
|
Wnt10a
|
wingless-type MMTV integration site family, member 10A |
| chr1_+_211582077 | 24.64 |
ENSRNOT00000023619
|
Pwwp2b
|
PWWP domain containing 2B |
| chr15_-_47442664 | 24.45 |
ENSRNOT00000072994
|
Prss55
|
protease, serine, 55 |
| chr1_+_101603222 | 24.35 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr4_+_159403501 | 24.15 |
ENSRNOT00000086525
|
Akap3
|
A-kinase anchoring protein 3 |
| chr14_-_9456990 | 24.04 |
ENSRNOT00000002918
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chrX_+_142344878 | 24.01 |
ENSRNOT00000056621
|
LOC108349258
|
high mobility group protein B4-like |
| chr10_+_57239993 | 23.97 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr4_+_70977556 | 23.91 |
ENSRNOT00000031984
|
LOC680112
|
hypothetical protein LOC680112 |
| chr1_-_89194602 | 23.83 |
ENSRNOT00000028518
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr9_+_93445002 | 22.86 |
ENSRNOT00000025029
|
LOC501180
|
similar to hypothetical protein MGC35154 |
| chr4_+_120156659 | 21.73 |
ENSRNOT00000050584
|
Dnajb8
|
DnaJ heat shock protein family (Hsp40) member B8 |
| chr10_-_56511583 | 21.49 |
ENSRNOT00000021402
|
LOC497940
|
similar to RIKEN cDNA 2810408A11 |
| chr19_+_49457677 | 21.03 |
ENSRNOT00000084445
|
Cenpn
|
centromere protein N |
| chr9_+_10172832 | 21.00 |
ENSRNOT00000074555
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr5_-_54482159 | 20.61 |
ENSRNOT00000050030
|
RGD1306195
|
similar to RAN protein |
| chr2_-_44504354 | 20.20 |
ENSRNOT00000013035
|
Ddx4
|
DEAD-box helicase 4 |
| chr15_+_80040842 | 19.98 |
ENSRNOT00000043065
|
RGD1306441
|
similar to RIKEN cDNA 4921530L21 |
| chr4_-_156945126 | 19.90 |
ENSRNOT00000064853
|
RGD1307916
|
LOC362433 |
| chr1_+_86938138 | 19.68 |
ENSRNOT00000075601
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr3_-_112371664 | 19.58 |
ENSRNOT00000079606
ENSRNOT00000012636 |
LOC100910478
|
leucine-rich repeat-containing protein 57-like |
| chr5_-_73494030 | 19.49 |
ENSRNOT00000022291
|
Actl7b
|
actin-like 7b |
| chr9_+_10535340 | 19.46 |
ENSRNOT00000075408
|
Znrf4
|
zinc and ring finger 4 |
| chr1_-_188245628 | 19.23 |
ENSRNOT00000022800
|
Tmc7
|
transmembrane channel-like 7 |
| chr4_-_88565292 | 18.65 |
ENSRNOT00000008948
|
Lancl2
|
LanC like 2 |
| chr2_-_246737997 | 18.02 |
ENSRNOT00000021719
|
Pdha2
|
pyruvate dehydrogenase (lipoamide) alpha 2 |
| chrX_+_13992064 | 17.80 |
ENSRNOT00000036543
|
LOC100363125
|
rCG42854-like |
| chr7_-_140951071 | 17.77 |
ENSRNOT00000082521
|
Fam186b
|
family with sequence similarity 186, member B |
| chr1_+_162768156 | 17.75 |
ENSRNOT00000049321
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr17_+_9746485 | 17.65 |
ENSRNOT00000072298
|
Pfn3
|
profilin 3 |
| chr14_+_39368530 | 17.33 |
ENSRNOT00000084367
|
Cox7b2
|
cytochrome c oxidase subunit 7B2 |
| chr2_+_127686925 | 17.20 |
ENSRNOT00000086653
ENSRNOT00000016946 |
Plk4
|
polo-like kinase 4 |
| chr9_-_14906410 | 17.19 |
ENSRNOT00000061674
|
LOC680955
|
hypothetical protein LOC680955 |
| chr5_-_113880911 | 17.14 |
ENSRNOT00000029441
|
Eqtn
|
equatorin |
| chr2_+_104290726 | 17.08 |
ENSRNOT00000017387
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr4_-_184096806 | 16.76 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chr18_+_25962855 | 16.66 |
ENSRNOT00000078822
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr10_-_45297385 | 16.55 |
ENSRNOT00000041187
|
Hist3h2bb
|
histone cluster 3 H2B family member b |
| chr4_+_174181644 | 16.30 |
ENSRNOT00000011555
|
Capza3
|
capping actin protein of muscle Z-line alpha subunit 3 |
| chr17_-_9695292 | 16.29 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr5_-_75116490 | 16.13 |
ENSRNOT00000042788
|
Txndc8
|
thioredoxin domain containing 8 |
| chr11_+_52828116 | 16.12 |
ENSRNOT00000035340
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr6_+_8669722 | 16.11 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr4_-_157078130 | 16.10 |
ENSRNOT00000015570
|
Clstn3
|
calsyntenin 3 |
| chr4_-_183697531 | 15.99 |
ENSRNOT00000055441
|
Amn1
|
antagonist of mitotic exit network 1 homolog |
| chr10_+_65454658 | 15.82 |
ENSRNOT00000037048
|
Proca1
|
protein interacting with cyclin A1 |
| chr17_+_85382116 | 15.04 |
ENSRNOT00000002513
|
Spag6
|
sperm associated antigen 6 |
| chr16_+_90613870 | 15.03 |
ENSRNOT00000079334
|
Shcbp1
|
SHC binding and spindle associated 1 |
| chr2_+_260039651 | 14.93 |
ENSRNOT00000073873
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr10_-_36716601 | 14.92 |
ENSRNOT00000038838
|
LOC497899
|
similar to hypothetical protein 4930503F14 |
| chr20_+_5262120 | 14.68 |
ENSRNOT00000000535
|
Brd2
|
bromodomain containing 2 |
| chr1_-_174411141 | 14.56 |
ENSRNOT00000065288
|
Nrip3
|
nuclear receptor interacting protein 3 |
| chr11_-_32469566 | 14.53 |
ENSRNOT00000002718
|
LOC498063
|
similar to RIKEN cDNA 4930563D23 |
| chr12_+_37984790 | 14.17 |
ENSRNOT00000001445
|
Vps37b
|
VPS37B, ESCRT-I subunit |
| chr5_-_141430659 | 14.12 |
ENSRNOT00000034944
|
Akirin1
|
akirin 1 |
| chr1_+_17602281 | 14.12 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr3_-_45210474 | 13.98 |
ENSRNOT00000091777
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr3_-_112385528 | 13.95 |
ENSRNOT00000049756
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr10_+_38104558 | 13.78 |
ENSRNOT00000067809
|
Fstl4
|
follistatin-like 4 |
| chr9_+_10760113 | 13.67 |
ENSRNOT00000073054
|
Arrdc5
|
arrestin domain containing 5 |
| chr10_-_74724472 | 13.65 |
ENSRNOT00000008846
|
Rad51c
|
RAD51 paralog C |
| chr10_-_91699424 | 13.55 |
ENSRNOT00000004650
|
Lyzl6
|
lysozyme-like 6 |
| chr17_-_43821536 | 13.46 |
ENSRNOT00000072286
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr3_+_125428260 | 13.37 |
ENSRNOT00000028892
|
Chgb
|
chromogranin B |
| chr19_+_55300395 | 13.14 |
ENSRNOT00000092169
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr3_-_160922341 | 13.13 |
ENSRNOT00000029206
|
Tp53tg5
|
TP53 target 5 |
| chr14_-_83062302 | 13.12 |
ENSRNOT00000086769
ENSRNOT00000085735 |
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr9_-_16095007 | 13.07 |
ENSRNOT00000073333
|
AABR07066782.1
|
|
| chr8_+_4085829 | 13.02 |
ENSRNOT00000031286
|
Actl9b
|
actin-like 9b |
| chr5_-_104920906 | 12.88 |
ENSRNOT00000071318
|
Fam154a
|
family with sequence similarity 154, member A |
| chr1_+_102849889 | 12.75 |
ENSRNOT00000066791
|
Gtf2h1
|
general transcription factor IIH subunit 1 |
| chr7_+_123168811 | 12.70 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr7_+_72924799 | 12.69 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr11_-_1437732 | 12.59 |
ENSRNOT00000037345
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr4_-_170740274 | 12.52 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chrX_-_79066932 | 12.39 |
ENSRNOT00000057460
|
RGD1561552
|
similar to WASP family 1 |
| chr5_-_39611053 | 12.35 |
ENSRNOT00000046595
|
Fhl5
|
four and a half LIM domains 5 |
| chr17_+_55008265 | 12.09 |
ENSRNOT00000033827
|
Zfp438
|
zinc finger protein 438 |
| chr1_+_142087208 | 12.05 |
ENSRNOT00000017532
|
Prc1
|
protein regulator of cytokinesis 1 |
| chr3_-_149356160 | 12.00 |
ENSRNOT00000037589
ENSRNOT00000081775 |
Sun5
|
Sad1 and UNC84 domain containing 5 |
| chr13_-_110257367 | 11.92 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr7_-_72328128 | 11.88 |
ENSRNOT00000008227
|
Tspyl5
|
TSPY-like 5 |
| chr9_-_114152716 | 11.75 |
ENSRNOT00000044153
|
AABR07068650.1
|
|
| chr2_-_137404996 | 11.69 |
ENSRNOT00000090907
|
LOC365791
|
similar to T-cell activation Rho GTPase-activating protein isoform b |
| chr9_-_114282799 | 11.45 |
ENSRNOT00000090539
|
LOC102555328
|
uncharacterized LOC102555328 |
| chr18_+_45023932 | 11.25 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr10_+_14519164 | 11.16 |
ENSRNOT00000079543
ENSRNOT00000046884 |
Ccdc154
|
coiled-coil domain containing 154 |
| chr7_+_140758615 | 11.12 |
ENSRNOT00000089448
|
Troap
|
trophinin associated protein |
| chr15_+_5904569 | 10.47 |
ENSRNOT00000072599
|
LOC102546376
|
disks large homolog 5-like |
| chr16_+_71241335 | 10.40 |
ENSRNOT00000066144
|
Letm2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr1_+_211205903 | 10.36 |
ENSRNOT00000023139
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr5_-_58163584 | 10.12 |
ENSRNOT00000060594
|
Ccl27
|
C-C motif chemokine ligand 27 |
| chr2_-_251970768 | 10.07 |
ENSRNOT00000020141
|
Wdr63
|
WD repeat domain 63 |
| chr13_-_104284068 | 9.97 |
ENSRNOT00000005407
|
LOC100912365
|
uncharacterized LOC100912365 |
| chr4_-_11497531 | 9.82 |
ENSRNOT00000078799
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr18_-_14016713 | 9.78 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr5_+_58937615 | 9.77 |
ENSRNOT00000080909
|
Tesk1
|
testis-specific kinase 1 |
| chr1_+_224938801 | 9.75 |
ENSRNOT00000073052
|
1700092M07Rik
|
RIKEN cDNA 1700092M07 gene |
| chr10_+_49368314 | 9.74 |
ENSRNOT00000004392
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr14_+_35683442 | 9.65 |
ENSRNOT00000003085
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr1_-_67302751 | 9.62 |
ENSRNOT00000041518
|
Vom1r42
|
vomeronasal 1 receptor 42 |
| chr15_-_33527031 | 9.54 |
ENSRNOT00000019979
|
Homez
|
homeobox and leucine zipper encoding |
| chr9_-_100253609 | 9.54 |
ENSRNOT00000036061
|
Kif1a
|
kinesin family member 1A |
| chr6_-_126622532 | 9.49 |
ENSRNOT00000038816
|
Moap1
|
modulator of apoptosis 1 |
| chr5_+_152721940 | 9.48 |
ENSRNOT00000039322
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr16_-_70998575 | 9.38 |
ENSRNOT00000019935
|
Kcnu1
|
potassium calcium-activated channel subfamily U member 1 |
| chrY_+_151937 | 9.35 |
ENSRNOT00000077988
ENSRNOT00000083382 |
Tspy1
|
testis specific protein, Y-linked 1 |
| chr2_-_112802073 | 9.18 |
ENSRNOT00000093081
|
Ect2
|
epithelial cell transforming 2 |
| chr14_+_7581189 | 9.15 |
ENSRNOT00000077215
|
LOC498330
|
similar to hypothetical protein MGC26744 |
| chr16_-_49522341 | 9.14 |
ENSRNOT00000081642
|
LOC100911140
|
coiled-coil domain-containing protein 110-like |
| chr10_+_59893067 | 9.11 |
ENSRNOT00000056443
|
Spata22
|
spermatogenesis associated 22 |
| chr7_+_27248115 | 9.08 |
ENSRNOT00000031946
ENSRNOT00000059545 |
LOC362863
|
first gene upstream of Nt5dc3 |
| chr17_-_89780691 | 9.07 |
ENSRNOT00000091830
|
LOC108348042
|
ankyrin repeat domain-containing protein 7-like |
| chr7_-_120558805 | 9.07 |
ENSRNOT00000087344
|
Pla2g6
|
phospholipase A2 group VI |
| chr12_+_504007 | 9.02 |
ENSRNOT00000001475
|
Brca2
|
BRCA2, DNA repair associated |
| chr20_+_13498926 | 8.97 |
ENSRNOT00000070992
ENSRNOT00000045375 |
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr19_-_55300403 | 8.96 |
ENSRNOT00000018591
|
Rnf166
|
ring finger protein 166 |
| chr16_-_32439421 | 8.83 |
ENSRNOT00000043100
|
Nek1
|
NIMA-related kinase 1 |
| chr9_-_82461903 | 8.83 |
ENSRNOT00000026654
|
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr10_+_39726507 | 8.66 |
ENSRNOT00000036822
|
Meikin
|
meiotic kinetochore factor |
| chr1_-_47213749 | 8.65 |
ENSRNOT00000024656
|
Dynlt1
|
dynein light chain Tctex-type 1 |
| chr12_-_13067341 | 8.62 |
ENSRNOT00000029692
|
Fam220a
|
family with sequence similarity 220, member A |
| chrX_+_16337102 | 8.60 |
ENSRNOT00000003954
|
Ccnb3
|
cyclin B3 |
| chr7_-_120559157 | 8.53 |
ENSRNOT00000090565
ENSRNOT00000016827 ENSRNOT00000017108 |
Pla2g6
|
phospholipase A2 group VI |
| chr4_+_157326727 | 8.50 |
ENSRNOT00000020493
|
Spsb2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr9_-_20219209 | 8.48 |
ENSRNOT00000072086
|
LOC100911548
|
SPRY domain-containing SOCS box protein 2-like |
| chr20_+_4967194 | 8.46 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr8_-_39830306 | 8.19 |
ENSRNOT00000040901
|
Ccdc15
|
coiled-coil domain containing 15 |
| chr2_-_123281856 | 7.98 |
ENSRNOT00000079745
|
Ccna2
|
cyclin A2 |
| chr7_-_22943159 | 7.90 |
ENSRNOT00000047547
|
Vom2r57
|
vomeronasal 2 receptor, 57 |
| chr8_-_62893087 | 7.89 |
ENSRNOT00000058490
|
Ccdc33
|
coiled-coil domain containing 33 |
| chr19_+_37668693 | 7.88 |
ENSRNOT00000084970
|
Pard6a
|
par-6 family cell polarity regulator alpha |
| chr7_-_19088972 | 7.87 |
ENSRNOT00000049266
|
Vom2r57
|
vomeronasal 2 receptor, 57 |
| chr7_-_82687130 | 7.86 |
ENSRNOT00000006791
|
Tmem74
|
transmembrane protein 74 |
| chr11_+_47200029 | 7.79 |
ENSRNOT00000079598
|
Nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr20_-_2088389 | 7.75 |
ENSRNOT00000090910
|
Znrd1as1
|
ZNRD1 antisense RNA 1 |
| chr1_+_72644333 | 7.72 |
ENSRNOT00000048976
ENSRNOT00000065051 |
Fam71e2
|
family with sequence similarity 71, member E2 |
| chr8_+_64364741 | 7.69 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chrX_+_43293551 | 7.67 |
ENSRNOT00000084562
|
LOC680190
|
hypothetical protein LOC680190 |
| chr15_+_27739251 | 7.55 |
ENSRNOT00000011840
|
Parp2
|
poly (ADP-ribose) polymerase 2 |
| chr7_-_30344426 | 7.50 |
ENSRNOT00000002638
ENSRNOT00000082016 |
Scyl2
|
SCY1 like pseudokinase 2 |
| chr20_-_37700937 | 7.44 |
ENSRNOT00000001051
|
Tbc1d32
|
TBC1 domain family, member 32 |
| chr16_+_70081035 | 7.41 |
ENSRNOT00000016098
|
LOC100912059
|
ubiquitin carboxyl-terminal hydrolase 12-like |
| chr2_+_86971147 | 7.35 |
ENSRNOT00000032909
|
Zfp458
|
zinc finger protein 458 |
| chr17_+_43617247 | 7.07 |
ENSRNOT00000075065
|
Hist1h2ao
|
histone cluster 1, H2ao |
| chr1_-_163071508 | 7.03 |
ENSRNOT00000019053
|
Myo7a
|
myosin VIIA |
| chr3_-_117389456 | 7.03 |
ENSRNOT00000007103
ENSRNOT00000081533 |
Myef2
|
myelin expression factor 2 |
| chrX_-_14783792 | 6.95 |
ENSRNOT00000087609
|
LOC691895
|
similar to ferritin, heavy polypeptide-like 17 |
| chr2_+_157453428 | 6.92 |
ENSRNOT00000051494
|
Lekr1
|
leucine, glutamate and lysine rich 1 |
| chr4_+_88834066 | 6.91 |
ENSRNOT00000009546
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr2_+_187302192 | 6.83 |
ENSRNOT00000015471
|
Isg20l2
|
interferon stimulated exonuclease gene 20-like 2 |
| chr16_-_32753278 | 6.83 |
ENSRNOT00000015759
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr20_-_10844178 | 6.79 |
ENSRNOT00000079207
|
Hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr8_+_114867062 | 6.73 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr15_+_90364605 | 6.68 |
ENSRNOT00000020876
|
Trim52
|
tripartite motif-containing 52 |
| chr8_+_122003916 | 6.58 |
ENSRNOT00000091980
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr7_-_44121130 | 6.53 |
ENSRNOT00000005706
|
Nts
|
neurotensin |
| chr20_-_3299420 | 6.45 |
ENSRNOT00000090999
|
Gnl1
|
G protein nucleolar 1 |
| chr18_+_62174670 | 6.43 |
ENSRNOT00000025362
|
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr6_-_135049728 | 6.26 |
ENSRNOT00000009556
|
Hsp90aa1
|
heat shock protein 90 alpha family class A member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf1_Creb5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 28.8 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 9.3 | 27.8 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 8.3 | 33.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 6.4 | 25.7 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 6.2 | 18.7 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 5.7 | 17.2 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 5.7 | 17.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 5.3 | 47.6 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 5.1 | 10.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 4.8 | 28.8 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 4.7 | 14.2 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 4.6 | 13.7 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 4.4 | 17.7 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 4.4 | 17.6 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 4.4 | 13.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 4.4 | 13.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 4.1 | 70.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 3.6 | 28.6 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 3.4 | 24.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 3.2 | 12.9 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 3.2 | 15.9 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 3.2 | 37.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 3.0 | 9.0 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 2.6 | 12.9 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 2.5 | 7.6 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 2.5 | 7.5 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 2.4 | 35.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 2.3 | 6.9 | GO:0019389 | glucuronoside metabolic process(GO:0019389) response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) |
| 2.2 | 8.7 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 2.0 | 21.7 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 2.0 | 29.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 1.8 | 5.5 | GO:0042441 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 1.7 | 24.9 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 1.6 | 9.8 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 1.6 | 16.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 1.5 | 9.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 1.5 | 12.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 1.5 | 4.4 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 1.3 | 22.7 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 1.3 | 6.5 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 1.3 | 16.7 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 1.3 | 5.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 1.3 | 26.3 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 1.2 | 20.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 1.2 | 30.4 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 1.2 | 10.4 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 1.1 | 3.4 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 1.1 | 3.3 | GO:0042196 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 1.1 | 1.1 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 1.1 | 13.8 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 1.0 | 6.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 1.0 | 9.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.0 | 6.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.9 | 3.8 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.9 | 13.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.9 | 7.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.9 | 2.6 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.9 | 11.9 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.8 | 14.3 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.8 | 5.0 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.8 | 65.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.8 | 3.9 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.8 | 7.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.8 | 4.7 | GO:0021764 | amygdala development(GO:0021764) |
| 0.7 | 9.7 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.7 | 9.9 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.7 | 14.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.7 | 4.2 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.7 | 2.7 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.7 | 4.8 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.7 | 27.9 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.7 | 2.7 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.7 | 2.6 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.7 | 16.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.6 | 4.4 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.6 | 3.7 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.6 | 1.8 | GO:0060816 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.6 | 11.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.6 | 8.8 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.6 | 4.0 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.6 | 6.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.6 | 10.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.6 | 48.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.6 | 6.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 2.2 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.5 | 4.3 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.5 | 4.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.5 | 2.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.5 | 1.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.5 | 2.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.5 | 4.8 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.5 | 2.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.5 | 1.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 5.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.4 | 1.6 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.4 | 2.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.4 | 38.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.4 | 1.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.4 | 3.0 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.4 | 2.2 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.4 | 3.6 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 54.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.3 | 12.5 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.3 | 1.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.3 | 3.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.3 | 1.3 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 2.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 2.9 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.3 | 3.8 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.3 | 4.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.3 | 2.0 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.3 | 7.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.3 | 2.8 | GO:1901679 | pyrimidine-containing compound transmembrane transport(GO:0072531) nucleotide transmembrane transport(GO:1901679) |
| 0.3 | 4.8 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.3 | 9.0 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 0.8 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.3 | 4.7 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.3 | 3.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 2.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 0.8 | GO:0021539 | subthalamus development(GO:0021539) pulmonary vein morphogenesis(GO:0060577) |
| 0.3 | 9.5 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.3 | 2.7 | GO:0032342 | aldosterone biosynthetic process(GO:0032342) |
| 0.3 | 7.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 1.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 2.6 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 30.0 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 0.7 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.2 | 0.6 | GO:0030186 | melatonin metabolic process(GO:0030186) negative regulation of male gonad development(GO:2000019) |
| 0.2 | 3.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.0 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.2 | 1.6 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.2 | 5.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 3.2 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.2 | 6.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 3.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 8.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.2 | 12.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.2 | 1.6 | GO:0016926 | protein desumoylation(GO:0016926) regulation of DNA endoreduplication(GO:0032875) |
| 0.2 | 16.0 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.2 | 4.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.6 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 2.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.4 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 1.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.8 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 2.5 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.1 | 0.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 2.3 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 5.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 5.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 5.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.6 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 2.1 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 1.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 4.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 4.2 | GO:0045429 | positive regulation of nitric oxide biosynthetic process(GO:0045429) |
| 0.1 | 0.8 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 5.7 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 6.6 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 0.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 1.6 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 2.2 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 0.2 | GO:0044827 | modulation by host of viral genome replication(GO:0044827) positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 2.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 15.7 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 2.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.9 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.3 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 4.0 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 3.0 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 2.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 5.2 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.1 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 2.0 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 2.5 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 121.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 7.1 | 35.3 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 4.3 | 13.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 4.3 | 17.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 3.4 | 17.2 | GO:0098536 | deuterosome(GO:0098536) |
| 3.4 | 20.2 | GO:0071547 | piP-body(GO:0071547) |
| 3.1 | 56.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 2.7 | 13.7 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) Holliday junction resolvase complex(GO:0048476) |
| 2.6 | 12.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 2.5 | 27.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 2.4 | 11.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 2.4 | 33.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 2.3 | 9.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 2.2 | 30.4 | GO:0031045 | dense core granule(GO:0031045) |
| 2.0 | 14.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 1.9 | 17.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 1.8 | 18.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 1.8 | 5.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.7 | 40.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 1.6 | 16.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 1.4 | 12.8 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 1.4 | 4.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.3 | 3.9 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 1.2 | 18.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 1.2 | 161.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 1.0 | 14.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.0 | 7.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 1.0 | 9.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 1.0 | 3.9 | GO:0097196 | Shu complex(GO:0097196) |
| 0.8 | 23.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.8 | 38.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.7 | 12.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.7 | 24.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.6 | 4.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.6 | 11.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.6 | 9.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.6 | 4.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.5 | 9.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.5 | 8.7 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.5 | 5.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.5 | 12.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.5 | 2.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.5 | 2.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.4 | 12.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.4 | 4.3 | GO:0000801 | central element(GO:0000801) |
| 0.4 | 3.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 1.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.4 | 1.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.3 | 26.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 1.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.3 | 5.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 2.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 5.5 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.3 | 7.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 2.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 18.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.3 | 4.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 1.8 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 17.0 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.3 | 4.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 4.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 12.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 2.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 4.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 1.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 1.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 4.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 5.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 7.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 1.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 4.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 3.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 20.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 2.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 2.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.8 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 4.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 2.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 9.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 2.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 4.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 8.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 8.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 5.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.4 | 65.6 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 9.1 | 72.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 8.0 | 24.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 5.6 | 16.8 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 4.5 | 18.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 3.2 | 28.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 3.0 | 9.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 2.9 | 8.6 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 2.8 | 13.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 2.7 | 24.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 2.6 | 25.7 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 2.5 | 9.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 2.3 | 9.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 2.3 | 13.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 2.2 | 28.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 2.2 | 17.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 2.1 | 25.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 1.8 | 5.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 1.7 | 21.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 1.6 | 4.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 1.5 | 4.4 | GO:0052723 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 1.4 | 16.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.3 | 18.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 1.3 | 10.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.1 | 3.4 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 1.1 | 29.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 1.1 | 4.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.0 | 13.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.0 | 11.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 1.0 | 2.0 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.9 | 7.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.9 | 5.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.9 | 177.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.9 | 9.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.9 | 6.3 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.9 | 2.7 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.9 | 6.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.8 | 7.6 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.8 | 4.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.8 | 6.9 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.8 | 11.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.8 | 12.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.8 | 4.5 | GO:0080084 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) 5S rDNA binding(GO:0080084) |
| 0.7 | 12.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.7 | 32.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.7 | 4.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.7 | 14.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.7 | 12.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.6 | 15.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.6 | 3.8 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.6 | 6.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 3.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.6 | 14.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.5 | 13.0 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.5 | 15.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.5 | 1.6 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.5 | 3.6 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.5 | 6.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.5 | 12.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.5 | 9.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.5 | 11.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 7.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 12.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.4 | 5.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 1.6 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.4 | 6.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 2.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.4 | 9.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.4 | 16.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.4 | 2.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.3 | 12.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.3 | 1.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.3 | 20.9 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.3 | 12.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 16.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.3 | 22.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.3 | 1.6 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 8.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.3 | 8.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 4.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 4.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 3.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 12.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 7.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 2.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 4.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 29.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.2 | 2.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 5.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 4.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 4.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 51.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 2.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 2.7 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 2.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 6.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 4.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 9.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 11.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 2.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 0.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 2.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 4.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 4.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 20.4 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.1 | 120.4 | GO:0003677 | DNA binding(GO:0003677) |
| 0.1 | 15.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 2.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 2.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 6.5 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 2.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.5 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.9 | 14.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.8 | 42.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.6 | 52.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.5 | 18.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.3 | 3.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.3 | 7.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 6.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.3 | 3.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.3 | 9.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 2.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.2 | 11.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 7.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 2.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 8.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 4.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 6.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 12.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 14.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 1.4 | 54.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.0 | 43.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.9 | 12.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.8 | 13.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.8 | 12.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.8 | 12.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.8 | 28.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.8 | 13.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.7 | 17.6 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.6 | 9.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 19.0 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.5 | 6.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.5 | 26.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.5 | 5.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 11.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.4 | 5.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 11.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.4 | 6.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 21.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.3 | 4.8 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.3 | 4.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.3 | 6.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.3 | 6.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 18.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.2 | 12.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 5.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 2.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 9.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 4.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 2.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 2.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 17.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 5.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 2.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 4.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 4.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 3.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 3.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 3.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 4.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 7.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |