Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
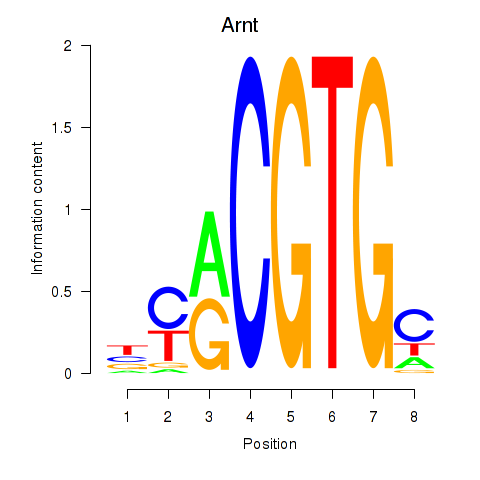
Results for Arnt
Z-value: 0.41
Transcription factors associated with Arnt
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arnt
|
ENSRNOG00000031174 | aryl hydrocarbon receptor nuclear translocator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arnt | rn6_v1_chr2_+_196594303_196594303 | -0.20 | 2.9e-04 | Click! |
Activity profile of Arnt motif
Sorted Z-values of Arnt motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_197991198 | 15.18 |
ENSRNOT00000056322
|
Ciart
|
circadian associated repressor of transcription |
| chr2_-_197991574 | 14.51 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr7_+_38945836 | 12.47 |
ENSRNOT00000006455
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr1_+_261229347 | 8.18 |
ENSRNOT00000018485
|
Ubtd1
|
ubiquitin domain containing 1 |
| chr2_-_44504354 | 7.38 |
ENSRNOT00000013035
|
Ddx4
|
DEAD-box helicase 4 |
| chr19_+_37226186 | 7.26 |
ENSRNOT00000075933
ENSRNOT00000065013 |
Hsf4
|
heat shock transcription factor 4 |
| chr3_+_72238981 | 6.92 |
ENSRNOT00000011006
|
Slc43a1
|
solute carrier family 43 member 1 |
| chr4_-_180234804 | 6.57 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr10_+_16542882 | 5.62 |
ENSRNOT00000028146
|
Stc2
|
stanniocalcin 2 |
| chr5_-_139227196 | 5.46 |
ENSRNOT00000050941
|
Foxo6
|
forkhead box O6 |
| chr9_-_113331319 | 3.84 |
ENSRNOT00000020681
|
Vapa
|
VAMP associated protein A |
| chr3_+_113415774 | 3.68 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr3_-_159802952 | 3.44 |
ENSRNOT00000011610
|
Oser1
|
oxidative stress responsive serine-rich 1 |
| chr1_-_93949187 | 3.41 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr2_+_189400696 | 3.41 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr4_+_95498003 | 3.29 |
ENSRNOT00000008358
|
Atoh1
|
atonal bHLH transcription factor 1 |
| chr7_+_123043503 | 3.24 |
ENSRNOT00000026258
ENSRNOT00000086355 |
Tef
|
TEF, PAR bZIP transcription factor |
| chr1_+_102849889 | 3.09 |
ENSRNOT00000066791
|
Gtf2h1
|
general transcription factor IIH subunit 1 |
| chr2_-_34185682 | 3.07 |
ENSRNOT00000066925
ENSRNOT00000082755 |
Nln
|
neurolysin |
| chr17_+_86199623 | 2.98 |
ENSRNOT00000022727
|
Ptf1a
|
pancreas specific transcription factor, 1a |
| chr2_+_115337439 | 2.98 |
ENSRNOT00000015779
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr8_-_36760742 | 2.97 |
ENSRNOT00000017307
|
Ddx25
|
DEAD-box helicase 25 |
| chr4_-_157679962 | 2.76 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr20_-_3401273 | 2.53 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr20_-_5533448 | 2.49 |
ENSRNOT00000000568
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr4_-_98593664 | 2.41 |
ENSRNOT00000007927
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr20_-_5533600 | 2.37 |
ENSRNOT00000072319
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr2_+_34186091 | 2.34 |
ENSRNOT00000016129
|
Sgtb
|
small glutamine rich tetratricopeptide repeat containing beta |
| chr14_-_35652709 | 2.32 |
ENSRNOT00000003080
|
Gsx2
|
GS homeobox 2 |
| chr9_+_81783349 | 2.26 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr1_-_86948845 | 2.17 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr7_-_75422268 | 1.98 |
ENSRNOT00000080218
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr2_-_189400323 | 1.93 |
ENSRNOT00000024364
|
Ubap2l
|
ubiquitin associated protein 2-like |
| chr8_+_22402890 | 1.86 |
ENSRNOT00000079230
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr1_-_234670113 | 1.68 |
ENSRNOT00000017133
|
LOC499331
|
similar to hypothetical protein D030056L22 |
| chr3_+_79678201 | 1.61 |
ENSRNOT00000087604
ENSRNOT00000079709 |
Mtch2
|
mitochondrial carrier 2 |
| chr8_-_67869019 | 1.55 |
ENSRNOT00000066009
|
Pias1
|
protein inhibitor of activated STAT, 1 |
| chr17_+_27496353 | 1.53 |
ENSRNOT00000077994
ENSRNOT00000019040 ENSRNOT00000089736 |
Ssr1
|
signal sequence receptor subunit 1 |
| chr15_-_34314345 | 1.48 |
ENSRNOT00000026552
|
Ipo4
|
importin 4 |
| chr7_-_18634079 | 1.47 |
ENSRNOT00000010031
|
Angptl4
|
angiopoietin-like 4 |
| chr4_-_82160240 | 1.45 |
ENSRNOT00000038775
ENSRNOT00000058985 |
Hoxa4
|
homeo box A4 |
| chr1_-_94404211 | 1.42 |
ENSRNOT00000019463
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr7_+_139762614 | 1.40 |
ENSRNOT00000031157
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr7_-_58219790 | 1.35 |
ENSRNOT00000067907
|
Tbc1d15
|
TBC1 domain family, member 15 |
| chr5_+_169506138 | 1.27 |
ENSRNOT00000014904
|
Rpl22
|
ribosomal protein L22 |
| chr3_+_164248731 | 1.26 |
ENSRNOT00000086459
|
Rnf114
|
ring finger protein 114 |
| chr1_+_77994203 | 1.25 |
ENSRNOT00000002044
|
Napa
|
NSF attachment protein alpha |
| chr3_-_173953684 | 1.23 |
ENSRNOT00000090468
|
Ppp1r3d
|
protein phosphatase 1, regulatory subunit 3D |
| chr15_-_27855999 | 1.13 |
ENSRNOT00000013225
|
Tmem55b
|
transmembrane protein 55B |
| chr4_-_82127051 | 1.06 |
ENSRNOT00000083658
ENSRNOT00000007807 |
Hoxa1
|
homeo box A1 |
| chr10_+_89236256 | 0.97 |
ENSRNOT00000027957
|
Psme3
|
proteasome activator subunit 3 |
| chr5_+_143500441 | 0.94 |
ENSRNOT00000045513
|
Grik3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr5_-_50362344 | 0.88 |
ENSRNOT00000035808
|
Zfp292
|
zinc finger protein 292 |
| chr1_+_220114228 | 0.79 |
ENSRNOT00000026718
|
Ctsf
|
cathepsin F |
| chr5_+_141572536 | 0.72 |
ENSRNOT00000023514
|
Rragc
|
Ras-related GTP binding C |
| chr12_+_21981755 | 0.71 |
ENSRNOT00000001871
|
LOC100910540
|
7SK snRNA methylphosphate capping enzyme-like |
| chr10_+_61746082 | 0.69 |
ENSRNOT00000003992
|
Tsr1
|
TSR1, ribosome maturation factor |
| chr11_-_60613718 | 0.62 |
ENSRNOT00000002906
|
Atg3
|
autophagy related 3 |
| chr1_+_12823363 | 0.54 |
ENSRNOT00000086790
|
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr1_-_129780356 | 0.51 |
ENSRNOT00000077479
|
Arrdc4
|
arrestin domain containing 4 |
| chr1_-_102849430 | 0.45 |
ENSRNOT00000086856
|
Saa4
|
serum amyloid A4 |
| chr19_-_55367353 | 0.38 |
ENSRNOT00000091139
|
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr4_+_117235023 | 0.27 |
ENSRNOT00000021030
|
Cct7
|
chaperonin containing TCP1 subunit 7 |
| chr13_-_51183269 | 0.26 |
ENSRNOT00000039540
|
Ppfia4
|
PTPRF interacting protein alpha 4 |
| chr16_+_7103998 | 0.26 |
ENSRNOT00000016581
|
Pbrm1
|
polybromo 1 |
| chr6_-_125853461 | 0.26 |
ENSRNOT00000007505
|
Atxn3
|
ataxin 3 |
| chr20_+_5455974 | 0.25 |
ENSRNOT00000000553
ENSRNOT00000092676 |
Pfdn6
|
prefoldin subunit 6 |
| chr8_+_50310405 | 0.22 |
ENSRNOT00000073507
|
Sik3
|
SIK family kinase 3 |
| chr4_-_82194927 | 0.07 |
ENSRNOT00000072302
|
LOC103692128
|
homeobox protein Hox-A9 |
| chr1_+_154377247 | 0.07 |
ENSRNOT00000092945
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr10_+_69737328 | 0.02 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr9_-_10734073 | 0.01 |
ENSRNOT00000071199
|
Kdm4b
|
lysine demethylase 4B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arnt
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 29.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 1.0 | 3.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.9 | 2.8 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.6 | 2.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.6 | 6.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.5 | 3.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.5 | 3.0 | GO:0006449 | regulation of translational termination(GO:0006449) positive regulation of translational elongation(GO:0045901) |
| 0.5 | 3.8 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.5 | 1.4 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.5 | 2.3 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.4 | 7.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.4 | 3.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 3.0 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.4 | 2.0 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 1.1 | GO:0021570 | rhombomere 3 development(GO:0021569) rhombomere 4 development(GO:0021570) |
| 0.4 | 5.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.3 | 7.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 1.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.3 | 1.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.3 | 2.3 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.2 | 2.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 6.9 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.2 | 3.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.5 | GO:0061428 | embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 1.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.9 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 3.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 5.5 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 0.4 | GO:0033625 | positive regulation of integrin activation(GO:0033625) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 1.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 1.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.7 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.3 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.7 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.0 | 0.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 3.4 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:1902961 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 1.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.5 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 1.4 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 3.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.4 | GO:0071547 | piP-body(GO:0071547) |
| 0.5 | 2.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 2.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 3.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 1.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 2.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 1.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 3.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 3.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 2.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 2.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.6 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0090544 | BAF-type complex(GO:0090544) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 1.0 | 3.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.9 | 6.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.7 | 2.8 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.6 | 30.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.4 | 3.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 1.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.4 | 1.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 1.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 6.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 0.6 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 3.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 2.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 10.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 1.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 2.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.7 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 8.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 4.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.5 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 2.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 2.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 7.3 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 5.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 3.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 3.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 4.5 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 7.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 6.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 3.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 3.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 2.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 3.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |