Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
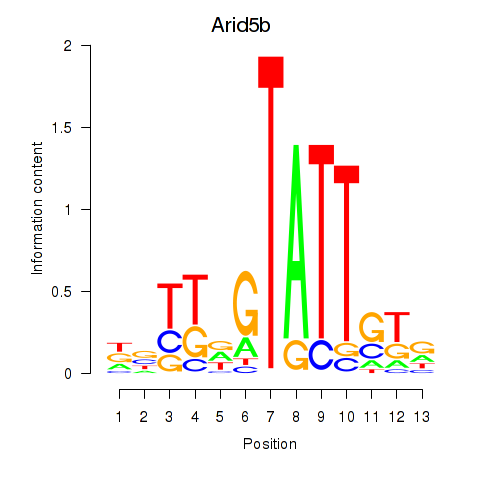
Results for Arid5b
Z-value: 0.63
Transcription factors associated with Arid5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid5b
|
ENSRNOG00000000635 | Arid5b |
Activity profile of Arid5b motif
Sorted Z-values of Arid5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_47721977 | 32.89 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr1_-_150395415 | 19.94 |
ENSRNOT00000018592
|
Folh1
|
folate hydrolase 1 |
| chr6_-_88917070 | 17.45 |
ENSRNOT00000000767
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr4_-_11497531 | 15.70 |
ENSRNOT00000078799
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr18_+_30574627 | 14.22 |
ENSRNOT00000060484
|
Pcdhb19
|
protocadherin beta 19 |
| chr3_+_117421604 | 13.54 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr10_-_59883839 | 13.39 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr2_+_121165137 | 13.01 |
ENSRNOT00000016236
|
Sox2
|
SRY box 2 |
| chr2_+_236625357 | 12.85 |
ENSRNOT00000081248
|
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr7_-_12874215 | 12.09 |
ENSRNOT00000011837
|
Hcn2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr3_-_46726946 | 11.77 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr6_-_43862131 | 11.70 |
ENSRNOT00000089859
|
Cys1
|
cystin 1 |
| chr4_+_140377565 | 11.60 |
ENSRNOT00000082723
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr14_-_66978499 | 11.37 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr2_+_83393282 | 10.59 |
ENSRNOT00000044871
|
Ctnnd2
|
catenin delta 2 |
| chr2_-_9023104 | 9.98 |
ENSRNOT00000039652
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr9_-_79898912 | 9.97 |
ENSRNOT00000022076
|
March4
|
membrane associated ring-CH-type finger 4 |
| chr6_-_42630983 | 9.58 |
ENSRNOT00000071977
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr1_-_100969560 | 9.29 |
ENSRNOT00000035908
|
Cpt1c
|
carnitine palmitoyltransferase 1c |
| chr2_-_197991198 | 9.28 |
ENSRNOT00000056322
|
Ciart
|
circadian associated repressor of transcription |
| chr2_-_26699333 | 8.56 |
ENSRNOT00000024459
|
Sv2c
|
synaptic vesicle glycoprotein 2c |
| chr10_-_66848388 | 8.55 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr18_-_58423196 | 8.51 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr20_+_3558827 | 8.26 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr4_+_61912210 | 8.18 |
ENSRNOT00000013569
|
Bpgm
|
bisphosphoglycerate mutase |
| chr1_+_153861948 | 8.10 |
ENSRNOT00000087067
|
Me3
|
malic enzyme 3 |
| chr18_+_29352749 | 7.91 |
ENSRNOT00000025137
|
Slc4a9
|
solute carrier family 4 member 9 |
| chr4_-_55398941 | 7.81 |
ENSRNOT00000075324
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr3_-_13525983 | 7.63 |
ENSRNOT00000082036
|
Pbx3
|
PBX homeobox 3 |
| chr1_-_157461588 | 7.18 |
ENSRNOT00000068402
|
Ankrd42
|
ankyrin repeat domain 42 |
| chr1_+_170205591 | 6.98 |
ENSRNOT00000071063
|
LOC686660
|
similar to olfactory receptor 692 |
| chr5_-_16140896 | 6.86 |
ENSRNOT00000029503
|
Xkr4
|
XK related 4 |
| chr5_-_173233188 | 6.52 |
ENSRNOT00000055343
|
Tmem88b
|
transmembrane protein 88B |
| chr1_-_43638161 | 6.32 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr10_-_62664466 | 6.24 |
ENSRNOT00000078730
|
Git1
|
GIT ArfGAP 1 |
| chr13_+_92264231 | 5.54 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr2_+_22950018 | 5.49 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chrX_+_111798233 | 5.45 |
ENSRNOT00000078582
|
Prps1
|
phosphoribosyl pyrophosphate synthetase 1 |
| chr6_-_2961510 | 5.31 |
ENSRNOT00000090688
|
Dhx57
|
DExH-box helicase 57 |
| chr2_+_186980992 | 5.22 |
ENSRNOT00000020717
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chrX_+_106253355 | 4.71 |
ENSRNOT00000004252
|
LOC102552920
|
armadillo repeat-containing X-linked protein 5-like |
| chr15_-_95514259 | 4.69 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr8_-_119889661 | 4.69 |
ENSRNOT00000011780
|
Stac
|
SH3 and cysteine rich domain |
| chr14_+_69800156 | 4.63 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr12_-_17311112 | 4.30 |
ENSRNOT00000001732
|
Gper1
|
G protein-coupled estrogen receptor 1 |
| chr19_-_54648227 | 4.13 |
ENSRNOT00000025424
|
Klhdc4
|
kelch domain containing 4 |
| chr10_-_1744647 | 4.12 |
ENSRNOT00000081886
|
AABR07028997.1
|
|
| chr10_-_52290657 | 4.09 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr5_-_70463546 | 3.47 |
ENSRNOT00000043184
|
AC118841.1
|
|
| chr18_+_14756684 | 3.34 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr4_-_165456677 | 3.03 |
ENSRNOT00000082207
|
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr4_-_28310178 | 2.98 |
ENSRNOT00000084021
|
RGD1563091
|
similar to OEF2 |
| chr8_-_50277797 | 2.84 |
ENSRNOT00000082508
|
Pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr2_+_186980793 | 2.83 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr10_-_64550145 | 2.81 |
ENSRNOT00000050232
|
Nxn
|
nucleoredoxin |
| chr10_-_34990943 | 2.74 |
ENSRNOT00000075555
|
Rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr20_+_4363508 | 2.73 |
ENSRNOT00000077205
|
Ager
|
advanced glycosylation end product-specific receptor |
| chr3_-_72081079 | 2.72 |
ENSRNOT00000007914
|
Tmx2
|
thioredoxin-related transmembrane protein 2 |
| chr5_+_58995249 | 2.70 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr18_-_81428971 | 2.63 |
ENSRNOT00000065201
|
Zfp407
|
zinc finger protein 407 |
| chr17_+_34704616 | 2.46 |
ENSRNOT00000090706
ENSRNOT00000083674 ENSRNOT00000077110 |
Exoc2
|
exocyst complex component 2 |
| chr8_+_18795525 | 2.34 |
ENSRNOT00000050430
|
Olr1124
|
olfactory receptor 1124 |
| chr4_-_67301102 | 2.21 |
ENSRNOT00000034549
|
Dennd2a
|
DENN domain containing 2A |
| chr3_+_75265525 | 2.12 |
ENSRNOT00000013319
|
Olr554
|
olfactory receptor 554 |
| chr11_+_74014983 | 2.11 |
ENSRNOT00000040884
|
NEWGENE_2724
|
glycoprotein V platelet |
| chr17_-_2705123 | 2.01 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr12_-_23624212 | 1.88 |
ENSRNOT00000064405
ENSRNOT00000001943 |
Rasa4
|
RAS p21 protein activator 4 |
| chr4_+_34237762 | 1.82 |
ENSRNOT00000084163
|
Mios
|
meiosis regulator for oocyte development |
| chr15_-_9086282 | 1.82 |
ENSRNOT00000008989
|
Thrb
|
thyroid hormone receptor beta |
| chr9_-_80295446 | 1.80 |
ENSRNOT00000023769
|
Tnp1
|
transition protein 1 |
| chr11_-_43992598 | 1.79 |
ENSRNOT00000002260
|
Cldnd1
|
claudin domain containing 1 |
| chr8_-_21481735 | 1.70 |
ENSRNOT00000038069
|
Zfp426
|
zinc finger protein 426 |
| chr4_+_123760743 | 1.69 |
ENSRNOT00000013498
|
Ccdc174
|
coiled-coil domain containing 174 |
| chr5_-_117612123 | 1.66 |
ENSRNOT00000065112
|
Dock7
|
dedicator of cytokinesis 7 |
| chr1_-_172552099 | 1.64 |
ENSRNOT00000090373
|
Olr257
|
olfactory receptor 257 |
| chr18_+_17550350 | 1.42 |
ENSRNOT00000078870
|
RGD1562608
|
similar to KIAA1328 protein |
| chr20_+_1647441 | 1.40 |
ENSRNOT00000000986
|
Olr1730
|
olfactory receptor 1730 |
| chr5_-_155709215 | 1.37 |
ENSRNOT00000018118
|
Cdc42
|
cell division cycle 42 |
| chrX_+_156210002 | 1.35 |
ENSRNOT00000077744
|
Olr1768
|
olfactory receptor 1768 |
| chr1_+_60117804 | 1.19 |
ENSRNOT00000080437
|
Vom1r8
|
vomeronasal 1 receptor 8 |
| chr1_+_167051209 | 1.18 |
ENSRNOT00000055249
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr4_+_129574264 | 1.15 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chr1_-_225948626 | 1.14 |
ENSRNOT00000077823
|
Incenp
|
inner centromere protein |
| chr5_+_135029955 | 1.11 |
ENSRNOT00000074860
|
LOC100911669
|
uncharacterized LOC100911669 |
| chr10_+_35872619 | 1.03 |
ENSRNOT00000059190
|
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr3_+_148448732 | 1.03 |
ENSRNOT00000083812
|
Ttll9
|
tubulin tyrosine ligase like 9 |
| chr8_+_117576788 | 0.98 |
ENSRNOT00000027584
|
Ip6k2
|
inositol hexakisphosphate kinase 2 |
| chr8_-_70436028 | 0.98 |
ENSRNOT00000077152
|
Slc24a1
|
solute carrier family 24 member 1 |
| chr18_+_60392376 | 0.80 |
ENSRNOT00000023890
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr4_+_44321883 | 0.79 |
ENSRNOT00000091095
|
Tes
|
testin LIM domain protein |
| chr7_+_132378273 | 0.79 |
ENSRNOT00000010990
|
LOC690142
|
hypothetical protein LOC690142 |
| chr17_+_45199178 | 0.78 |
ENSRNOT00000080047
|
Zscan26
|
zinc finger and SCAN domain containing 26 |
| chr10_+_71322145 | 0.54 |
ENSRNOT00000079418
|
Synrg
|
synergin, gamma |
| chr3_-_73124644 | 0.53 |
ENSRNOT00000012711
|
Olr455
|
olfactory receptor 455 |
| chr1_-_60407295 | 0.49 |
ENSRNOT00000078350
|
Vom1r12
|
vomeronasal 1 receptor 12 |
| chr4_+_88048267 | 0.39 |
ENSRNOT00000044913
|
Vom1r81
|
vomeronasal 1 receptor 81 |
| chr2_+_226825635 | 0.36 |
ENSRNOT00000042173
|
AABR07013200.1
|
|
| chr3_+_102841615 | 0.28 |
ENSRNOT00000075178
|
Olr770
|
olfactory receptor 770 |
| chr3_-_76626605 | 0.18 |
ENSRNOT00000051292
|
Olr625
|
olfactory receptor 625 |
| chr11_+_81757983 | 0.13 |
ENSRNOT00000088503
|
Tbccd1
|
TBCC domain containing 1 |
| chr2_-_258932200 | 0.08 |
ENSRNOT00000045905
|
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr15_-_39742103 | 0.00 |
ENSRNOT00000074587
ENSRNOT00000036781 ENSRNOT00000071919 |
Setdb2
|
SET domain, bifurcated 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arid5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.5 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 3.8 | 11.4 | GO:0071623 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 3.3 | 19.9 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 3.3 | 13.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 2.4 | 12.1 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 2.4 | 11.8 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 2.0 | 15.7 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 1.8 | 12.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.8 | 5.4 | GO:0034418 | urate biosynthetic process(GO:0034418) |
| 1.7 | 11.6 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 1.4 | 4.3 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 1.4 | 10.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 1.2 | 8.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.0 | 4.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.9 | 4.7 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.9 | 2.7 | GO:0072714 | response to selenite ion(GO:0072714) negative regulation of connective tissue replacement(GO:1905204) |
| 0.8 | 8.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.8 | 13.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.7 | 8.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.6 | 13.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.6 | 9.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.6 | 7.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.5 | 1.4 | GO:0071338 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.4 | 7.6 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.4 | 3.0 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.4 | 1.7 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.4 | 17.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.4 | 9.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.4 | 2.5 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.3 | 6.2 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.3 | 1.8 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.3 | 9.6 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 8.6 | GO:0048679 | regulation of collateral sprouting(GO:0048670) regulation of axon regeneration(GO:0048679) |
| 0.3 | 1.8 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 7.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.2 | 0.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 5.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.2 | 8.5 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.2 | 10.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 4.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 14.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 5.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 11.7 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 1.2 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 1.9 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 | 8.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 1.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 2.8 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 1.0 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.0 | 1.0 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 1.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 2.9 | 11.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 1.9 | 9.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 1.7 | 10.0 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 1.6 | 15.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 1.1 | 5.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.7 | 2.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.6 | 5.5 | GO:0008091 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) cuticular plate(GO:0032437) |
| 0.4 | 1.2 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.4 | 4.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 11.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 9.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 1.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 1.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 17.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 5.5 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 4.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 3.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 8.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.1 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 7.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 13.0 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 10.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 1.8 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 8.6 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 18.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 4.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 10.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 15.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 1.0 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.1 | 1.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 7.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 8.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 20.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 7.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 28.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.5 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 4.5 | 13.4 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 4.3 | 12.8 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 3.9 | 15.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 2.9 | 11.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 2.8 | 11.4 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 2.3 | 9.3 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 2.0 | 8.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 2.0 | 7.8 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 1.4 | 19.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 1.4 | 8.1 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.2 | 12.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.2 | 8.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.1 | 5.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.1 | 9.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.9 | 8.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.8 | 13.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.7 | 2.8 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.7 | 5.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.5 | 2.7 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.5 | 1.4 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.4 | 4.3 | GO:1990239 | estrogen receptor activity(GO:0030284) steroid hormone binding(GO:1990239) |
| 0.3 | 4.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 1.0 | GO:0052723 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.3 | 7.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 2.8 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 9.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 7.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 1.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 5.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 2.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 10.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 11.7 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 10.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 11.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 8.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 5.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 8.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 12.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 7.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.4 | 11.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.4 | 6.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 13.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.3 | 11.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 6.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 9.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 8.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 12.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 11.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.6 | 15.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.4 | 9.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.4 | 7.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 9.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 2.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 5.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 4.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 11.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 12.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 8.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 10.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 5.4 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |