Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Arid5a
Z-value: 0.21
Transcription factors associated with Arid5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid5a
|
ENSRNOG00000015382 | AT-rich interaction domain 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arid5a | rn6_v1_chr9_+_42871950_42871950 | -0.33 | 8.8e-10 | Click! |
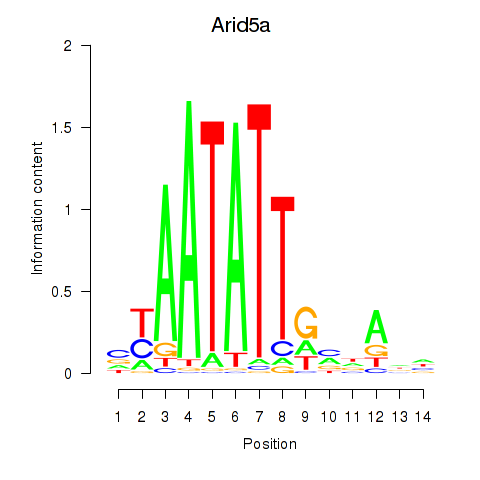
Activity profile of Arid5a motif
Sorted Z-values of Arid5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_170431073 | 9.54 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chrX_-_23144324 | 5.94 |
ENSRNOT00000000178
ENSRNOT00000081239 |
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr13_+_27465930 | 3.39 |
ENSRNOT00000003314
|
Serpinb10
|
serpin family B member 10 |
| chr9_-_32019205 | 3.07 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chrX_+_40460047 | 2.88 |
ENSRNOT00000010970
|
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr3_-_48831417 | 1.38 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr20_+_17750744 | 1.36 |
ENSRNOT00000000745
|
RGD1559903
|
similar to RIKEN cDNA 1700049L16 |
| chr14_-_41786084 | 1.35 |
ENSRNOT00000059439
|
Grxcr1
|
glutaredoxin and cysteine rich domain containing 1 |
| chr3_+_70327193 | 1.24 |
ENSRNOT00000089165
|
Fsip2
|
fibrous sheath-interacting protein 2 |
| chr13_-_90443157 | 0.94 |
ENSRNOT00000006862
|
Nhlh1
|
nescient helix loop helix 1 |
| chrX_-_122062799 | 0.86 |
ENSRNOT00000086852
|
Gm4907
|
predicted gene 4907 |
| chr17_+_43734461 | 0.17 |
ENSRNOT00000072564
|
Hist1h1d
|
histone cluster 1, H1d |
| chr6_+_123034304 | 0.06 |
ENSRNOT00000078938
|
Cpg1
|
candidate plasticity gene 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arid5a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.5 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 1.0 | 2.9 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.7 | 5.9 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.6 | 3.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 1.3 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 3.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 9.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 5.9 | GO:0061695 | transferase complex, transferring phosphorus-containing groups(GO:0061695) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.8 | 5.9 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |