Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
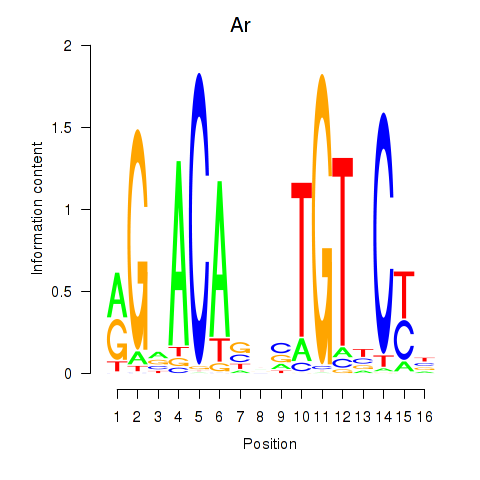
Results for Ar
Z-value: 0.91
Transcription factors associated with Ar
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ar
|
ENSRNOG00000005639 | androgen receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ar | rn6_v1_chrX_+_67656253_67656253 | -0.03 | 6.1e-01 | Click! |
Activity profile of Ar motif
Sorted Z-values of Ar motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_138483612 | 35.84 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr15_-_5509839 | 24.76 |
ENSRNOT00000080252
ENSRNOT00000081979 ENSRNOT00000085137 ENSRNOT00000049746 |
Spetex-2F
|
Spetex-2F protein |
| chr10_+_14519164 | 24.30 |
ENSRNOT00000079543
ENSRNOT00000046884 |
Ccdc154
|
coiled-coil domain containing 154 |
| chr4_-_98735346 | 23.76 |
ENSRNOT00000008473
|
Tex37
|
testis expressed 37 |
| chr13_+_51218468 | 22.70 |
ENSRNOT00000033636
|
LOC289035
|
similar to UDP-N-acetylglucosamine:a-1,3-D-mannoside beta-1,4-N-acetylgluco |
| chr10_+_103992309 | 21.54 |
ENSRNOT00000065292
|
Trim80
|
tripartite motif protein 80 |
| chr6_-_127630734 | 21.51 |
ENSRNOT00000089931
|
Serpina1
|
serpin family A member 1 |
| chr17_+_72160735 | 21.17 |
ENSRNOT00000038817
|
Itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_4953879 | 21.13 |
ENSRNOT00000003455
|
Tnp2
|
transition protein 2 |
| chr9_-_16095007 | 21.10 |
ENSRNOT00000073333
|
AABR07066782.1
|
|
| chr7_-_26144466 | 20.69 |
ENSRNOT00000009162
|
Fhl4
|
four and a half LIM domains 4 |
| chr6_+_76349362 | 20.24 |
ENSRNOT00000043224
|
Aldoart2
|
aldolase 1 A retrogene 2 |
| chr10_-_56429748 | 19.87 |
ENSRNOT00000020675
ENSRNOT00000092704 |
Spem1
|
spermatid maturation 1 |
| chr2_-_243475639 | 19.34 |
ENSRNOT00000089222
|
RGD1309170
|
similar to hypothetical protein DKFZp434G072 |
| chr5_-_58309627 | 19.28 |
ENSRNOT00000031509
ENSRNOT00000085709 |
Fam205c
|
family with sequence similarity 205, member C |
| chr1_-_189238776 | 18.70 |
ENSRNOT00000020817
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr6_-_47848026 | 18.16 |
ENSRNOT00000011048
ENSRNOT00000090017 |
Allc
|
allantoicase |
| chr16_+_29674793 | 17.83 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr15_-_5467047 | 17.70 |
ENSRNOT00000042743
|
Spetex-2F
|
Spetex-2F protein |
| chr1_-_162533893 | 16.91 |
ENSRNOT00000016783
|
Aamdc
|
adipogenesis associated, Mth938 domain containing |
| chr5_-_122828398 | 16.56 |
ENSRNOT00000030112
|
RGD1562532
|
hypothetical gene supported by BC079057 |
| chr13_+_91080341 | 16.55 |
ENSRNOT00000000058
|
Crp
|
C-reactive protein |
| chr10_+_49368314 | 16.24 |
ENSRNOT00000004392
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr17_-_8619737 | 15.87 |
ENSRNOT00000065217
|
RGD1562024
|
RGD1562024 |
| chr3_+_143129248 | 15.79 |
ENSRNOT00000006667
|
Cst8
|
cystatin 8 |
| chr5_-_127273656 | 15.41 |
ENSRNOT00000057341
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr10_+_4950484 | 15.40 |
ENSRNOT00000003450
|
Prm2
|
protamine 2 |
| chr7_-_100382897 | 15.22 |
ENSRNOT00000006510
|
LOC500876
|
LOC500876 |
| chr5_+_124442293 | 15.06 |
ENSRNOT00000041922
|
RGD1564074
|
similar to novel protein |
| chr6_-_47811853 | 14.85 |
ENSRNOT00000010942
|
Allc
|
allantoicase |
| chr15_-_42751330 | 14.67 |
ENSRNOT00000066478
|
Adam2
|
ADAM metallopeptidase domain 2 |
| chr5_+_136117858 | 14.59 |
ENSRNOT00000087764
|
Tmem53
|
transmembrane protein 53 |
| chr6_-_26624092 | 14.54 |
ENSRNOT00000008113
|
Trim54
|
tripartite motif-containing 54 |
| chr17_+_56935451 | 14.51 |
ENSRNOT00000058966
|
RGD1564129
|
similar to hypothetical protein 4930474N05 |
| chr16_+_12976906 | 14.44 |
ENSRNOT00000029573
|
LOC685463
|
similar to Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS) |
| chr13_+_109646455 | 14.41 |
ENSRNOT00000073985
|
LOC498316
|
hypothetical LOC498316 |
| chr16_-_12742010 | 14.32 |
ENSRNOT00000077513
|
LOC100911649
|
ral guanine nucleotide dissociation stimulator-like |
| chr15_+_67092304 | 14.30 |
ENSRNOT00000040244
|
AABR07018646.1
|
|
| chr5_-_27180819 | 14.18 |
ENSRNOT00000045483
|
AABR07047293.1
|
|
| chr6_+_110749705 | 14.00 |
ENSRNOT00000084348
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr4_-_6062641 | 13.95 |
ENSRNOT00000074846
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr5_-_73494030 | 13.93 |
ENSRNOT00000022291
|
Actl7b
|
actin-like 7b |
| chr7_+_2689501 | 13.88 |
ENSRNOT00000041341
|
Apof
|
apolipoprotein F |
| chr6_-_23543172 | 13.86 |
ENSRNOT00000006073
|
Spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr3_+_151609602 | 13.81 |
ENSRNOT00000065052
|
Spag4
|
sperm associated antigen 4 |
| chr2_+_104290726 | 13.76 |
ENSRNOT00000017387
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr1_+_201337416 | 13.61 |
ENSRNOT00000067742
|
Btbd16
|
BTB domain containing 16 |
| chr3_+_152143811 | 13.51 |
ENSRNOT00000026578
|
LOC100911109
|
sperm-associated antigen 4 protein-like |
| chr1_-_206394346 | 13.26 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chr1_+_18353812 | 13.25 |
ENSRNOT00000029394
|
AABR07000581.1
|
|
| chr1_-_154507039 | 13.16 |
ENSRNOT00000078883
|
Ccdc83
|
coiled-coil domain containing 83 |
| chr1_+_267618565 | 13.03 |
ENSRNOT00000076251
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr15_-_5417570 | 12.97 |
ENSRNOT00000061525
|
Spetex-2F
|
Spetex-2F protein |
| chr20_-_25826658 | 12.91 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr10_-_107376645 | 12.59 |
ENSRNOT00000046213
|
Cep295nl
|
CEP295 N-terminal like |
| chr5_-_108747666 | 12.41 |
ENSRNOT00000072732
|
Zfp353
|
zinc finger protein 353 |
| chr10_-_43892604 | 12.30 |
ENSRNOT00000003801
|
LOC691286
|
similar to RIKEN cDNA 4930504O13 |
| chr17_+_45175121 | 11.99 |
ENSRNOT00000080417
|
Nkapl
|
NFKB activating protein-like |
| chr1_-_143398093 | 11.97 |
ENSRNOT00000078916
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr10_+_96639924 | 11.73 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chrX_-_97262013 | 11.70 |
ENSRNOT00000040168
|
Cldn34c4
|
claudin 34C4 |
| chr16_-_12662336 | 11.66 |
ENSRNOT00000080161
|
RGD1559508
|
similar to hypothetical protein 4930474N05 |
| chr13_+_75177965 | 11.63 |
ENSRNOT00000007321
|
Sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chrX_+_709306 | 11.46 |
ENSRNOT00000071047
|
Ssx2
|
synovial sarcoma, X breakpoint 2 |
| chr13_-_74005486 | 11.35 |
ENSRNOT00000090173
|
AABR07021465.1
|
|
| chr14_+_94776233 | 11.22 |
ENSRNOT00000070839
|
AABR07016166.1
|
|
| chr1_+_83043760 | 11.19 |
ENSRNOT00000074720
|
LOC103689938
|
carcinoembryonic antigen-related cell adhesion molecule 1-like |
| chr3_-_72219246 | 11.15 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr3_-_51054378 | 11.14 |
ENSRNOT00000089243
|
Grb14
|
growth factor receptor bound protein 14 |
| chr2_-_216443518 | 11.01 |
ENSRNOT00000022496
|
Amy1a
|
amylase, alpha 1A |
| chr6_-_27323992 | 10.96 |
ENSRNOT00000013198
|
RGD1559683
|
similar to RIKEN cDNA 1700001C02 |
| chr19_-_26082719 | 10.95 |
ENSRNOT00000083159
|
Rnaseh2a
|
ribonuclease H2, subunit A |
| chr6_+_128073344 | 10.80 |
ENSRNOT00000014073
|
LOC500712
|
Ab1-233 |
| chr8_-_132910905 | 10.59 |
ENSRNOT00000008630
|
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chrX_-_106576314 | 10.37 |
ENSRNOT00000037823
|
Nxf3
|
nuclear RNA export factor 3 |
| chr11_-_36533073 | 10.33 |
ENSRNOT00000033486
|
Lca5l
|
LCA5L, lebercilin like |
| chr1_-_205630073 | 10.31 |
ENSRNOT00000037064
|
Tex36
|
testis expressed 36 |
| chr11_+_64488194 | 10.29 |
ENSRNOT00000030268
|
RGD1306995
|
similar to hypothetical protein FLJ32859 |
| chr3_+_170475831 | 10.28 |
ENSRNOT00000006949
|
Fam209a
|
family with sequence similarity 209, member A |
| chr9_+_95309759 | 10.25 |
ENSRNOT00000051438
|
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr20_+_5170215 | 10.06 |
ENSRNOT00000001139
|
Ncr3
|
natural cytotoxicity triggering receptor 3 |
| chr6_-_127816055 | 10.03 |
ENSRNOT00000013175
|
Serpina3m
|
serine (or cysteine) proteinase inhibitor, clade A, member 3M |
| chr6_+_26780352 | 10.00 |
ENSRNOT00000009916
|
Prr30
|
proline rich 30 |
| chrX_+_62366453 | 9.98 |
ENSRNOT00000089828
|
Arx
|
aristaless related homeobox |
| chr10_-_56426012 | 9.86 |
ENSRNOT00000092506
ENSRNOT00000084068 |
LOC497938
|
similar to RIKEN cDNA 4933402P03 |
| chr10_-_5511955 | 9.85 |
ENSRNOT00000039429
|
Atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr3_-_148073554 | 9.80 |
ENSRNOT00000055407
|
Defb27
|
defensin beta 27 |
| chr15_-_28314459 | 9.72 |
ENSRNOT00000042055
ENSRNOT00000040540 |
Ndrg2
|
NDRG family member 2 |
| chr2_-_208633945 | 9.68 |
ENSRNOT00000049155
|
Pifo
|
primary cilia formation |
| chr12_+_45319501 | 9.66 |
ENSRNOT00000090630
ENSRNOT00000041732 |
RGD1561114
|
similar to hypothetical protein 4930474N05 |
| chr20_-_32115310 | 9.66 |
ENSRNOT00000067883
ENSRNOT00000081916 |
Vps26a
|
VPS26 retromer complex component A |
| chr4_+_1470716 | 9.64 |
ENSRNOT00000044223
|
Olr1235
|
olfactory receptor 1235 |
| chrX_-_71681073 | 9.55 |
ENSRNOT00000037212
|
LOC100361265
|
rCG64283-like |
| chr9_-_27447877 | 9.53 |
ENSRNOT00000085195
|
Gsta1
|
glutathione S-transferase alpha 1 |
| chr5_+_58752879 | 9.44 |
ENSRNOT00000071201
|
Atp8b5p
|
ATPase, class I, type 8B, member 5, pseudogene |
| chr4_-_69149520 | 9.41 |
ENSRNOT00000017611
|
Prss58
|
protease, serine, 58 |
| chr11_-_57993548 | 9.28 |
ENSRNOT00000002957
|
Nectin3
|
nectin cell adhesion molecule 3 |
| chr20_-_4542947 | 9.26 |
ENSRNOT00000086140
|
Cfb
|
complement factor B |
| chr4_+_57823411 | 9.24 |
ENSRNOT00000030462
ENSRNOT00000088235 |
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr6_-_135026449 | 9.10 |
ENSRNOT00000009444
|
RGD1305298
|
hypothetical LOC299330 |
| chr1_+_81372650 | 9.02 |
ENSRNOT00000088829
|
Zfp428
|
zinc finger protein 428 |
| chr15_+_108453147 | 8.98 |
ENSRNOT00000018486
|
Timm8a2
|
translocase of inner mitochondrial membrane 8A2 |
| chr4_-_178168690 | 8.95 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chr8_+_5893249 | 8.81 |
ENSRNOT00000014041
|
Mmp7
|
matrix metallopeptidase 7 |
| chr7_+_141953794 | 8.62 |
ENSRNOT00000041938
|
Tmprss12
|
transmembrane protease, serine 12 |
| chr7_-_80796670 | 8.50 |
ENSRNOT00000010539
|
Abra
|
actin-binding Rho activating protein |
| chr3_-_2719513 | 8.48 |
ENSRNOT00000020997
|
Lcn12
|
lipocalin 12 |
| chr3_+_154986059 | 8.41 |
ENSRNOT00000082528
|
LOC100911217
|
adipogenin-like |
| chr16_+_6970342 | 8.40 |
ENSRNOT00000061294
ENSRNOT00000048344 |
Itih4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr9_+_27402381 | 8.40 |
ENSRNOT00000077372
|
Gsta3
|
glutathione S-transferase alpha 3 |
| chr1_-_18511695 | 8.37 |
ENSRNOT00000075402
|
AABR07000583.1
|
|
| chr8_-_72841496 | 8.32 |
ENSRNOT00000057641
ENSRNOT00000040808 ENSRNOT00000085894 ENSRNOT00000024575 ENSRNOT00000048044 ENSRNOT00000024493 |
Tpm1
|
tropomyosin 1, alpha |
| chr19_+_50246402 | 8.32 |
ENSRNOT00000018795
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr8_-_107490093 | 8.12 |
ENSRNOT00000046832
|
LOC684466
|
similar to Fas apoptotic inhibitory molecule 1 (rFAIM) |
| chr14_+_26662965 | 8.11 |
ENSRNOT00000002621
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr7_+_12941994 | 8.07 |
ENSRNOT00000010832
|
Odf3l2
|
outer dense fiber of sperm tails 3-like 2 |
| chr3_-_161027943 | 8.02 |
ENSRNOT00000078633
|
Spint3
|
serine peptidase inhibitor, Kunitz type, 3 |
| chr7_-_117267803 | 7.97 |
ENSRNOT00000082271
|
Plec
|
plectin |
| chr7_-_117267402 | 7.70 |
ENSRNOT00000088945
|
Plec
|
plectin |
| chr13_-_85695421 | 7.64 |
ENSRNOT00000005768
|
Lrrc52
|
leucine rich repeat containing 52 |
| chr13_+_70157522 | 7.58 |
ENSRNOT00000036906
|
Apobec4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chr13_+_52889737 | 7.56 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr7_-_117259141 | 7.55 |
ENSRNOT00000040762
|
Plec
|
plectin |
| chr1_+_260051406 | 7.51 |
ENSRNOT00000079734
|
AABR07072054.1
|
|
| chr4_-_100099517 | 7.50 |
ENSRNOT00000014277
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr1_+_142034423 | 7.49 |
ENSRNOT00000090006
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr7_-_12518684 | 7.46 |
ENSRNOT00000018691
ENSRNOT00000093426 |
Gpx4
|
glutathione peroxidase 4 |
| chr1_-_198104109 | 7.43 |
ENSRNOT00000026186
|
Sult1a1
|
sulfotransferase family 1A member 1 |
| chr1_-_19522171 | 7.36 |
ENSRNOT00000045386
|
AABR07000595.1
|
|
| chr4_-_6046477 | 7.33 |
ENSRNOT00000073925
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr19_+_15094309 | 7.27 |
ENSRNOT00000083500
|
Ces1f
|
carboxylesterase 1F |
| chr3_+_143109316 | 7.17 |
ENSRNOT00000006555
|
LOC257643
|
cystatin SC |
| chr20_-_13667333 | 7.12 |
ENSRNOT00000031472
|
LOC103694872
|
coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial |
| chr13_+_83073866 | 7.03 |
ENSRNOT00000075996
|
Dpt
|
dermatopontin |
| chrX_+_72684329 | 7.01 |
ENSRNOT00000057644
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chr3_+_2438089 | 6.97 |
ENSRNOT00000013335
|
Fam166a
|
family with sequence similarity 166, member A |
| chr2_+_127770676 | 6.93 |
ENSRNOT00000068405
|
Abhd18
|
abhydrolase domain containing 18 |
| chr7_-_117259791 | 6.81 |
ENSRNOT00000086550
|
Plec
|
plectin |
| chr1_-_88723973 | 6.68 |
ENSRNOT00000082555
|
NEWGENE_1306714
|
WD repeat domain 62-like 1 |
| chr9_+_41006800 | 6.65 |
ENSRNOT00000079413
ENSRNOT00000034214 |
Prss39
|
protease, serine, 39 |
| chr19_-_11302938 | 6.64 |
ENSRNOT00000038212
|
AC128848.1
|
|
| chr2_-_184951950 | 6.61 |
ENSRNOT00000093486
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr2_+_178117466 | 6.58 |
ENSRNOT00000065115
ENSRNOT00000084198 |
LOC499643
|
similar to hypothetical protein FLJ25371 |
| chr13_-_51373399 | 6.52 |
ENSRNOT00000060656
|
Mgat4e
|
MGAT4 family, member E |
| chr4_-_181348799 | 6.47 |
ENSRNOT00000033743
|
LOC690784
|
hypothetical protein LOC690783 |
| chr9_+_94228960 | 6.41 |
ENSRNOT00000083137
ENSRNOT00000080897 |
Akp3
|
alkaline phosphatase 3, intestine, not Mn requiring |
| chr9_-_113900190 | 6.38 |
ENSRNOT00000016965
|
Ndufv2
|
NADH:ubiquinone oxidoreductase core subunit V2 |
| chr20_+_34633157 | 6.34 |
ENSRNOT00000000469
|
Pln
|
phospholamban |
| chr15_+_18332910 | 6.28 |
ENSRNOT00000085822
|
Fam3d
|
family with sequence similarity 3, member D |
| chr5_-_164585634 | 6.27 |
ENSRNOT00000065985
|
Mfn2
|
mitofusin 2 |
| chr16_-_8350067 | 6.20 |
ENSRNOT00000026932
|
LOC100362432
|
mitochondrial import inner membrane translocase subunit Tim23-like |
| chr1_+_221443896 | 6.18 |
ENSRNOT00000073942
|
Tmem262
|
transmembrane protein 262 |
| chr5_+_154112076 | 6.15 |
ENSRNOT00000044969
|
Myom3
|
myomesin 3 |
| chr13_-_52834134 | 6.12 |
ENSRNOT00000038874
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr12_+_45026886 | 6.09 |
ENSRNOT00000001500
|
Pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr7_-_117719605 | 6.03 |
ENSRNOT00000077172
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr5_+_172825072 | 6.01 |
ENSRNOT00000068525
|
Cfap74
|
cilia and flagella associated protein 74 |
| chr1_+_89008117 | 5.94 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr16_-_49318383 | 5.92 |
ENSRNOT00000045721
|
Cfap97
|
cilia and flagella associated protein 97 |
| chr10_-_11942885 | 5.90 |
ENSRNOT00000061019
|
Olr1356
|
olfactory receptor 1356 |
| chr7_-_117268300 | 5.82 |
ENSRNOT00000091840
|
Plec
|
plectin |
| chr16_+_75083172 | 5.65 |
ENSRNOT00000058002
|
Defb37
|
defensin beta 37 |
| chr1_-_216953999 | 5.62 |
ENSRNOT00000028126
|
Mrgprg
|
MAS related GPR family member G |
| chr2_-_185168476 | 5.59 |
ENSRNOT00000093447
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr12_-_31249118 | 5.57 |
ENSRNOT00000058586
ENSRNOT00000073105 |
Adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr3_-_80543031 | 5.57 |
ENSRNOT00000022233
|
F2
|
coagulation factor II |
| chr2_+_104020955 | 5.53 |
ENSRNOT00000045586
|
Mtfr1
|
mitochondrial fission regulator 1 |
| chr20_-_30947484 | 5.52 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr20_+_3364814 | 5.45 |
ENSRNOT00000001077
|
RGD1302996
|
hypothetical protein MGC:15854 |
| chr10_-_75120247 | 5.43 |
ENSRNOT00000011402
|
Lpo
|
lactoperoxidase |
| chr2_-_3124543 | 5.39 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr16_+_50181316 | 5.35 |
ENSRNOT00000077662
|
F11
|
coagulation factor XI |
| chr4_+_148649223 | 5.35 |
ENSRNOT00000040801
|
Olr829
|
olfactory receptor 829 |
| chr5_+_58393233 | 5.32 |
ENSRNOT00000000142
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr6_+_26546924 | 5.30 |
ENSRNOT00000007763
|
Eif2b4
|
eukaryotic translation initiation factor 2B subunit delta |
| chr13_+_99335020 | 5.29 |
ENSRNOT00000029787
|
AABR07021930.1
|
|
| chr7_+_143754892 | 5.25 |
ENSRNOT00000085896
|
Soat2
|
sterol O-acyltransferase 2 |
| chr1_-_94610854 | 5.20 |
ENSRNOT00000020383
|
Plekhf1
|
pleckstrin homology and FYVE domain containing 1 |
| chr5_-_110326940 | 5.15 |
ENSRNOT00000049470
|
Zfp352
|
zinc finger protein 352 |
| chr1_+_282567674 | 5.14 |
ENSRNOT00000090543
|
Ces2i
|
carboxylesterase 2I |
| chr16_-_7681576 | 5.09 |
ENSRNOT00000026550
|
Colq
|
collagen like tail subunit of asymmetric acetylcholinesterase |
| chr1_-_216942782 | 5.08 |
ENSRNOT00000049057
|
RGD1562011
|
similar to MAS-related G-protein coupled receptor, member G |
| chr3_+_2182957 | 5.08 |
ENSRNOT00000011633
|
Pnpla7
|
patatin-like phospholipase domain containing 7 |
| chr2_-_119537837 | 5.05 |
ENSRNOT00000015200
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr4_-_115453659 | 4.98 |
ENSRNOT00000065847
|
Tex261
|
testis expressed 261 |
| chr8_+_56411911 | 4.93 |
ENSRNOT00000039083
|
LOC100359687
|
mitochondrial ribosomal protein L1-like |
| chr4_-_176606382 | 4.93 |
ENSRNOT00000065576
|
Recql
|
RecQ like helicase |
| chr1_-_170694872 | 4.83 |
ENSRNOT00000075443
|
LOC100911839
|
olfactory receptor 2D3-like |
| chr4_-_27726943 | 4.83 |
ENSRNOT00000068499
|
LOC500007
|
similar to EF hand calcium binding domain 1 |
| chr5_+_58393603 | 4.79 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr10_-_62699723 | 4.79 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr1_+_171274305 | 4.76 |
ENSRNOT00000051038
|
Olr234
|
olfactory receptor 234 |
| chr1_+_203160323 | 4.75 |
ENSRNOT00000027919
|
AABR07005844.1
|
|
| chr3_-_94419048 | 4.71 |
ENSRNOT00000015775
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr6_-_38453797 | 4.71 |
ENSRNOT00000009100
|
Ddx1
|
DEAD-box helicase 1 |
| chr15_-_29856947 | 4.63 |
ENSRNOT00000066990
|
AABR07017662.2
|
|
| chr10_+_35857797 | 4.59 |
ENSRNOT00000004517
|
Cby3
|
chibby family member 3 |
| chr15_+_108286453 | 4.55 |
ENSRNOT00000016986
|
Ubac2
|
UBA domain containing 2 |
| chr8_+_85951191 | 4.53 |
ENSRNOT00000037665
|
Cd109
|
CD109 molecule |
| chr1_-_212022212 | 4.51 |
ENSRNOT00000074298
|
Cfap46
|
cilia and flagella associated protein 46 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ar
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.0 | 33.0 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 7.2 | 21.5 | GO:0033986 | response to methanol(GO:0033986) |
| 3.4 | 10.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 3.0 | 21.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 2.9 | 8.8 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 2.9 | 11.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 2.8 | 8.4 | GO:1901377 | toxin catabolic process(GO:0009407) mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) secondary metabolite catabolic process(GO:0090487) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 2.8 | 8.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 2.4 | 9.5 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 2.2 | 11.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 1.9 | 35.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.9 | 5.6 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) response to thyrotropin-releasing hormone(GO:1905225) |
| 1.8 | 10.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 1.7 | 13.8 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 1.7 | 10.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.6 | 12.9 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 1.5 | 8.9 | GO:2000741 | regulation of timing of neuron differentiation(GO:0060164) positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 1.4 | 13.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 1.4 | 4.3 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 1.4 | 4.2 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 1.4 | 4.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.3 | 18.7 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 1.3 | 3.9 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.3 | 3.9 | GO:1904753 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 1.3 | 11.6 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 1.3 | 16.6 | GO:0010988 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 1.2 | 3.6 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 1.2 | 2.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 1.2 | 9.3 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 1.1 | 5.5 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 1.1 | 7.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 1.1 | 3.2 | GO:0006106 | fumarate metabolic process(GO:0006106) glycerol biosynthetic process(GO:0006114) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 1.1 | 7.4 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 1.0 | 11.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.0 | 5.9 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.9 | 8.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.9 | 5.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.9 | 4.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.9 | 9.5 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.8 | 5.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.8 | 4.9 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.8 | 2.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.8 | 2.4 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.8 | 3.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.8 | 2.4 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.8 | 6.3 | GO:1990910 | response to hypobaric hypoxia(GO:1990910) |
| 0.8 | 5.4 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.8 | 4.5 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.7 | 10.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.7 | 2.9 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.7 | 5.0 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.7 | 22.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.7 | 2.7 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.7 | 6.1 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.7 | 4.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.7 | 0.7 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.7 | 5.3 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.6 | 17.2 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.6 | 3.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.6 | 1.8 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.6 | 21.3 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.5 | 2.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.5 | 3.7 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.5 | 8.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.5 | 5.6 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.5 | 1.4 | GO:0048850 | cuticle development(GO:0042335) hypophysis morphogenesis(GO:0048850) |
| 0.4 | 4.9 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.4 | 1.8 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.4 | 1.3 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.4 | 4.6 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.4 | 11.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.4 | 9.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.4 | 3.2 | GO:0035878 | nail development(GO:0035878) |
| 0.4 | 1.9 | GO:0042262 | dGTP catabolic process(GO:0006203) DNA protection(GO:0042262) |
| 0.4 | 4.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.4 | 1.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.4 | 7.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.4 | 14.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.4 | 1.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.4 | 1.5 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.4 | 14.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.4 | 4.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.4 | 11.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.3 | 1.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.3 | 1.7 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.3 | 9.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.3 | 6.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 4.8 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.3 | 4.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.3 | 4.3 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.3 | 0.9 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.3 | 10.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.3 | 2.1 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.3 | 0.9 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.3 | 1.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.3 | 1.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.3 | 0.8 | GO:0000958 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.3 | 15.1 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.3 | 20.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.3 | 1.0 | GO:0019471 | glyoxylate catabolic process(GO:0009436) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.3 | 2.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.3 | 2.5 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.3 | 6.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 1.7 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.2 | 1.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 19.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.2 | 11.1 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.2 | 3.9 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 2.4 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.2 | 4.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 6.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 8.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.2 | 1.6 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.2 | 1.0 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.2 | 0.9 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 15.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.2 | 6.4 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.2 | 1.4 | GO:0009650 | UV protection(GO:0009650) |
| 0.2 | 7.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 2.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 0.7 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.2 | 0.8 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 5.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.4 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 2.0 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 0.1 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.1 | 1.4 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.4 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 2.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.2 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.1 | 1.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 4.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 6.0 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 2.5 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.1 | 1.9 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 1.2 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.7 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 11.0 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 11.1 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 5.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 1.0 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 4.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 0.5 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.9 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 1.3 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.1 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 1.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 2.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 3.9 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.1 | 5.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.1 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.1 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 2.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 1.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 69.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.6 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 1.8 | GO:1901031 | regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 3.5 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 1.0 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 2.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.8 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 1.3 | GO:0071806 | protein transmembrane transport(GO:0071806) |
| 0.0 | 0.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 16.2 | GO:0005975 | carbohydrate metabolic process(GO:0005975) |
| 0.0 | 1.1 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.4 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 1.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 3.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.6 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 4.4 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 11.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 2.4 | 9.7 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 2.3 | 31.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 2.1 | 6.3 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
| 2.0 | 11.7 | GO:0042627 | chylomicron(GO:0042627) |
| 1.7 | 35.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 1.2 | 8.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.2 | 13.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.9 | 5.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.9 | 21.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.9 | 9.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.6 | 6.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.6 | 4.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.6 | 2.4 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.5 | 4.9 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.5 | 17.3 | GO:0031430 | M band(GO:0031430) |
| 0.4 | 4.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.4 | 3.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.4 | 2.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 1.2 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.4 | 9.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 2.3 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.3 | 38.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 4.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 3.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 0.8 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.3 | 1.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 35.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 12.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 4.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 9.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 0.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 0.7 | GO:0070110 | interleukin-6 receptor complex(GO:0005896) ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.2 | 18.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 2.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 5.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 1.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 3.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 23.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.2 | 30.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.1 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.1 | 5.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 6.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 2.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.0 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 7.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 10.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 1.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 3.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 3.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 5.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 4.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 2.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 4.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 8.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 4.5 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 4.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 3.7 | 11.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 3.3 | 13.0 | GO:0030614 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 2.9 | 20.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.5 | 7.4 | GO:0050656 | steroid sulfotransferase activity(GO:0050294) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 2.4 | 16.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.3 | 18.7 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 2.3 | 29.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 2.2 | 11.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.4 | 12.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.2 | 8.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 1.1 | 3.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.1 | 31.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 1.0 | 5.0 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 1.0 | 35.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 1.0 | 3.9 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.9 | 7.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.9 | 6.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.8 | 4.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.8 | 4.0 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.8 | 6.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.8 | 2.4 | GO:0052871 | leukotriene-B4 20-monooxygenase activity(GO:0050051) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.7 | 10.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.7 | 2.1 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 0.7 | 4.2 | GO:0004144 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.7 | 5.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.7 | 3.5 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.7 | 6.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.7 | 77.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.6 | 1.9 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.6 | 1.9 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.6 | 11.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.6 | 3.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.6 | 2.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 4.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.5 | 1.6 | GO:0098603 | selenol Se-methyltransferase activity(GO:0098603) |
| 0.5 | 2.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.5 | 8.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.5 | 9.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.5 | 5.5 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.5 | 2.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.5 | 1.4 | GO:0016414 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 0.4 | 5.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.4 | 2.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.4 | 3.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.4 | 9.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.4 | 1.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.4 | 2.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.4 | 4.9 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 5.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 1.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.4 | 4.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.4 | 7.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 1.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.3 | 7.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 1.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.3 | 1.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.3 | 2.5 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.3 | 8.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.3 | 1.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 4.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.3 | 10.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.3 | 3.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.3 | 6.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 1.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.3 | 8.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 3.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 5.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 3.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 3.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 16.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 16.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 1.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 5.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 5.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 1.9 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 2.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 4.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.7 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.2 | 9.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 2.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 7.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 5.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 1.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 15.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 17.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.3 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 1.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 3.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.7 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 21.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 1.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 17.7 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.1 | 2.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 1.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 1.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 2.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 67.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 53.4 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 4.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 1.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 8.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.1 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.8 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 1.3 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.8 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 14.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.4 | 19.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 60.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 6.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 11.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 9.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 4.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 3.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 5.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 4.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 11.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.0 | 31.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 1.0 | 11.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.0 | 9.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.9 | 27.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 7.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 5.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.5 | 11.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.5 | 18.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.4 | 4.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 5.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 8.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 8.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 2.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 3.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 12.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 7.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 2.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 2.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 12.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 3.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 4.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |