Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Alx4
Z-value: 0.96
Transcription factors associated with Alx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Alx4
|
ENSRNOG00000000008 | ALX homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Alx4 | rn6_v1_chr3_+_82548959_82548959 | 0.29 | 1.1e-07 | Click! |
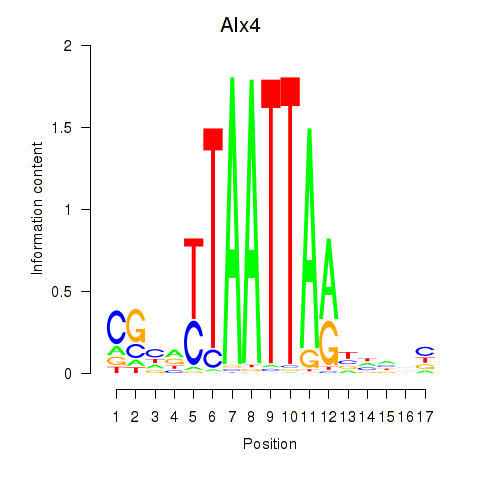
Activity profile of Alx4 motif
Sorted Z-values of Alx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_33606124 | 36.81 |
ENSRNOT00000065210
|
AC115371.1
|
|
| chr2_+_248398917 | 31.06 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr5_-_161981441 | 26.25 |
ENSRNOT00000020316
|
Pdpn
|
podoplanin |
| chr5_-_17061837 | 23.66 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr8_-_84506328 | 22.86 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr4_-_129619142 | 22.18 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chr8_-_78096302 | 20.99 |
ENSRNOT00000086185
ENSRNOT00000077999 |
Myzap
|
myocardial zonula adherens protein |
| chr15_+_87722221 | 20.94 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr4_+_144382945 | 20.91 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr5_-_17061361 | 20.89 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr17_+_23661429 | 20.42 |
ENSRNOT00000046523
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr11_+_36555416 | 19.68 |
ENSRNOT00000064981
|
Sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr4_+_155321553 | 19.64 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr12_-_5685448 | 17.91 |
ENSRNOT00000077167
|
Fry
|
FRY microtubule binding protein |
| chr10_+_69412017 | 17.82 |
ENSRNOT00000009448
|
Ccl2
|
C-C motif chemokine ligand 2 |
| chr17_+_63635086 | 17.21 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr2_+_219598162 | 16.78 |
ENSRNOT00000020297
|
Lrrc39
|
leucine rich repeat containing 39 |
| chr5_+_64476317 | 16.37 |
ENSRNOT00000017217
|
LOC108348074
|
collagen alpha-1(XV) chain-like |
| chr14_-_77810147 | 16.15 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr13_-_84331905 | 15.59 |
ENSRNOT00000004965
|
Dusp27
|
dual specificity phosphatase 27 (putative) |
| chr4_-_157304653 | 15.19 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr19_+_27404712 | 14.90 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr2_-_96509424 | 14.76 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr18_+_55666027 | 14.66 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr9_+_20241062 | 14.61 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr2_-_158156444 | 13.79 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr17_-_84247038 | 13.73 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr4_+_180291389 | 13.45 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr1_+_8310577 | 13.30 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr16_+_29674793 | 12.98 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr4_+_158088505 | 12.83 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr15_-_95514259 | 12.67 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr1_-_215033460 | 12.24 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr13_-_82005741 | 11.84 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr13_-_82006005 | 11.83 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr18_-_26656879 | 11.73 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr13_+_82369493 | 11.67 |
ENSRNOT00000003733
|
Sell
|
selectin L |
| chr13_+_87986240 | 11.47 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr2_+_58724855 | 11.47 |
ENSRNOT00000089609
|
Capsl
|
calcyphosine-like |
| chr4_-_41212072 | 11.33 |
ENSRNOT00000085596
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr1_-_278042312 | 11.32 |
ENSRNOT00000018693
|
Ablim1
|
actin-binding LIM protein 1 |
| chrX_-_14972675 | 11.06 |
ENSRNOT00000079664
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr12_-_31249118 | 10.79 |
ENSRNOT00000058586
ENSRNOT00000073105 |
Adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr4_-_109532234 | 10.78 |
ENSRNOT00000008665
|
Reg3g
|
regenerating family member 3 gamma |
| chr3_+_2591331 | 10.77 |
ENSRNOT00000017466
|
Sapcd2
|
suppressor APC domain containing 2 |
| chr18_-_55891710 | 10.71 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr13_-_91735361 | 10.67 |
ENSRNOT00000058090
|
Fcer1a
|
Fc fragment of IgE receptor Ia |
| chr5_-_168734296 | 10.50 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr16_-_29936307 | 10.50 |
ENSRNOT00000088707
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr3_+_20303979 | 10.50 |
ENSRNOT00000058331
|
AABR07051730.1
|
|
| chr11_+_24266345 | 10.47 |
ENSRNOT00000046973
|
Jam2
|
junctional adhesion molecule 2 |
| chr4_+_29535852 | 10.43 |
ENSRNOT00000087619
|
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr8_+_100260049 | 10.40 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr8_-_49280901 | 10.22 |
ENSRNOT00000021390
|
Cd3g
|
CD3g molecule |
| chr16_+_2537248 | 10.18 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr2_-_147392062 | 10.18 |
ENSRNOT00000021535
|
Tm4sf1
|
transmembrane 4 L six family member 1 |
| chr15_-_93765498 | 10.15 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr12_-_40590361 | 10.11 |
ENSRNOT00000067503
|
Tmem116
|
transmembrane protein 116 |
| chr18_-_61057365 | 10.07 |
ENSRNOT00000042352
|
Alpk2
|
alpha-kinase 2 |
| chr10_-_88266210 | 9.99 |
ENSRNOT00000090702
ENSRNOT00000020603 |
Hap1
|
huntingtin-associated protein 1 |
| chrX_+_84064427 | 9.97 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr18_-_3676188 | 9.88 |
ENSRNOT00000073811
|
Ankrd29
|
ankyrin repeat domain 29 |
| chr6_-_108920767 | 9.87 |
ENSRNOT00000006704
|
Prox2
|
prospero homeobox 2 |
| chr8_-_84835060 | 9.82 |
ENSRNOT00000007867
|
Lrrc1
|
leucine rich repeat containing 1 |
| chr2_+_145174876 | 9.52 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr17_+_32973695 | 9.51 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr10_-_51778939 | 9.48 |
ENSRNOT00000078675
ENSRNOT00000057562 |
Myocd
|
myocardin |
| chr8_+_2604962 | 9.47 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chr5_-_7941822 | 9.47 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr14_-_115052450 | 9.46 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr4_+_88184956 | 9.40 |
ENSRNOT00000077129
|
Vom1r83
|
vomeronasal 1 receptor 83 |
| chr5_+_152680407 | 9.32 |
ENSRNOT00000076864
|
Stmn1
|
stathmin 1 |
| chr13_-_50916982 | 9.11 |
ENSRNOT00000004408
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr4_+_101687327 | 9.10 |
ENSRNOT00000082501
|
AABR07060957.1
|
|
| chr7_+_144865608 | 9.03 |
ENSRNOT00000091596
ENSRNOT00000055285 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr14_+_113867209 | 8.92 |
ENSRNOT00000067591
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr11_-_17684903 | 8.66 |
ENSRNOT00000051213
|
Tmprss15
|
transmembrane protease, serine 15 |
| chr18_+_3887419 | 8.64 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr1_+_15834779 | 8.58 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr17_-_87826421 | 8.56 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr5_-_134927235 | 8.46 |
ENSRNOT00000016751
|
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chrX_+_96863891 | 8.43 |
ENSRNOT00000085665
|
AABR07040284.1
|
|
| chr12_+_41486076 | 8.37 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr10_-_67401836 | 8.37 |
ENSRNOT00000073071
|
Crlf3
|
cytokine receptor-like factor 3 |
| chr8_-_39460844 | 8.36 |
ENSRNOT00000048875
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr1_-_267463694 | 8.20 |
ENSRNOT00000084851
ENSRNOT00000016645 |
Col17a1
|
collagen type XVII alpha 1 chain |
| chr20_-_5020150 | 8.16 |
ENSRNOT00000001146
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr3_+_17889972 | 8.14 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr9_+_64745051 | 8.11 |
ENSRNOT00000021527
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr2_-_191294374 | 8.04 |
ENSRNOT00000067469
|
RGD1562234
|
similar to S100 calcium-binding protein, ventral prostate |
| chr5_-_22765429 | 7.99 |
ENSRNOT00000079432
|
Asph
|
aspartate-beta-hydroxylase |
| chrX_-_77675487 | 7.98 |
ENSRNOT00000042799
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chrX_+_40460047 | 7.94 |
ENSRNOT00000010970
|
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr6_-_51498337 | 7.90 |
ENSRNOT00000012487
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr4_+_169161585 | 7.83 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr6_-_108076186 | 7.79 |
ENSRNOT00000014814
|
Fam161b
|
family with sequence similarity 161, member B |
| chr6_+_76349362 | 7.74 |
ENSRNOT00000043224
|
Aldoart2
|
aldolase 1 A retrogene 2 |
| chr11_+_36851038 | 7.70 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr3_+_11763706 | 7.69 |
ENSRNOT00000078366
|
Sh2d3c
|
SH2 domain containing 3C |
| chr14_-_34561696 | 7.55 |
ENSRNOT00000059763
|
Srd5a3
|
steroid 5 alpha-reductase 3 |
| chr1_-_17378047 | 7.53 |
ENSRNOT00000020102
|
Themis
|
thymocyte selection associated |
| chr5_-_22769907 | 7.52 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr7_+_15785410 | 7.51 |
ENSRNOT00000082664
ENSRNOT00000073235 |
Zfp955a
|
zinc finger protein 955A |
| chr18_+_30890869 | 7.40 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr10_-_105668593 | 7.33 |
ENSRNOT00000016622
|
St6galnac2
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr4_+_169147243 | 7.25 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr12_-_23727535 | 7.23 |
ENSRNOT00000085911
ENSRNOT00000001950 |
Dtx2
|
deltex E3 ubiquitin ligase 2 |
| chr5_+_165724027 | 7.18 |
ENSRNOT00000018000
|
Casz1
|
castor zinc finger 1 |
| chr4_-_163463718 | 7.12 |
ENSRNOT00000085671
|
Klrc1
|
killer cell lectin like receptor C1 |
| chr1_+_101554642 | 7.07 |
ENSRNOT00000028474
|
Bcat2
|
branched chain amino acid transaminase 2 |
| chr4_+_31229913 | 7.06 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr10_-_74679858 | 7.05 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr6_+_95205153 | 6.99 |
ENSRNOT00000007339
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr17_+_44794130 | 6.99 |
ENSRNOT00000077571
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr10_-_89130339 | 6.98 |
ENSRNOT00000027640
|
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr20_+_3246739 | 6.92 |
ENSRNOT00000061299
|
RT1-T24-2
|
RT1 class I, locus T24, gene 2 |
| chr10_+_46722109 | 6.85 |
ENSRNOT00000032685
|
Drc3
|
dynein regulatory complex subunit 3 |
| chr17_-_54714914 | 6.83 |
ENSRNOT00000024336
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr7_+_2752680 | 6.80 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chrX_+_131381134 | 6.79 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr2_+_266315036 | 6.78 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr8_-_21492801 | 6.77 |
ENSRNOT00000077465
|
Zfp426
|
zinc finger protein 426 |
| chr9_+_71915421 | 6.75 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr4_+_69386698 | 6.67 |
ENSRNOT00000091655
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr6_-_21112734 | 6.58 |
ENSRNOT00000079819
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chrY_+_506149 | 6.52 |
ENSRNOT00000086056
|
Kdm5d
|
lysine demethylase 5D |
| chr6_+_91532467 | 6.51 |
ENSRNOT00000082500
ENSRNOT00000064668 |
Klhdc1
|
kelch domain containing 1 |
| chr17_-_86657473 | 6.48 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chrX_+_78042859 | 6.48 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr9_-_78368777 | 6.46 |
ENSRNOT00000020414
|
Bard1
|
BRCA1 associated RING domain 1 |
| chr2_-_170301348 | 6.45 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr8_-_18762922 | 6.41 |
ENSRNOT00000008423
|
Olr1122
|
olfactory receptor 1122 |
| chr1_-_63684189 | 6.41 |
ENSRNOT00000085651
|
Lilrc2
|
leukocyte immunoglobulin-like receptor, subfamily C, member 2 |
| chr17_-_15519282 | 6.32 |
ENSRNOT00000093577
|
Aspnl1
|
asporin-like 1 |
| chr20_-_10013190 | 6.24 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chr12_+_47590154 | 6.21 |
ENSRNOT00000045946
|
Git2
|
GIT ArfGAP 2 |
| chr2_-_182846061 | 6.21 |
ENSRNOT00000013025
|
Tlr2
|
toll-like receptor 2 |
| chrX_-_121731543 | 6.21 |
ENSRNOT00000018788
|
Klhl13
|
kelch-like family member 13 |
| chr10_-_896938 | 6.20 |
ENSRNOT00000086392
|
Nde1
|
nudE neurodevelopment protein 1 |
| chr17_-_43776460 | 6.19 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr15_+_4240203 | 6.12 |
ENSRNOT00000010178
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr1_+_227892956 | 6.08 |
ENSRNOT00000028483
|
AABR07006278.1
|
|
| chr5_-_162751128 | 6.04 |
ENSRNOT00000068281
|
RGD1559644
|
similar to novel protein similar to esterases |
| chr2_+_57276919 | 6.04 |
ENSRNOT00000063899
|
RGD1310081
|
similar to hypothetical protein FLJ13231 |
| chr7_-_15073052 | 6.01 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chr20_+_42966140 | 5.85 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr18_+_68408890 | 5.81 |
ENSRNOT00000039702
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr16_+_84465656 | 5.79 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr14_+_7171613 | 5.78 |
ENSRNOT00000081600
|
Klhl8
|
kelch-like family member 8 |
| chr10_+_64174931 | 5.76 |
ENSRNOT00000035948
|
RGD1565611
|
RGD1565611 |
| chr5_+_165465095 | 5.75 |
ENSRNOT00000031978
|
LOC691354
|
hypothetical protein LOC691354 |
| chr14_+_82769642 | 5.74 |
ENSRNOT00000065393
|
Ctbp1
|
C-terminal binding protein 1 |
| chr10_-_104075777 | 5.73 |
ENSRNOT00000004862
|
Hn1
|
hematological and neurological expressed 1 |
| chr4_+_98481520 | 5.71 |
ENSRNOT00000078381
ENSRNOT00000048493 |
AABR07060886.1
|
|
| chr13_-_44540516 | 5.71 |
ENSRNOT00000005244
|
Map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr16_-_18643309 | 5.69 |
ENSRNOT00000031680
|
Dydc2
|
DPY30 domain containing 2 |
| chr15_-_88670349 | 5.64 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr18_+_55466373 | 5.60 |
ENSRNOT00000074629
|
LOC102555392
|
interferon-inducible GTPase 1-like |
| chr8_-_17525906 | 5.57 |
ENSRNOT00000007855
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr5_-_99033107 | 5.57 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr7_-_5818625 | 5.55 |
ENSRNOT00000051699
|
Olr886
|
olfactory receptor 886 |
| chr2_-_158156150 | 5.53 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr16_-_3765917 | 5.52 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr20_+_3155652 | 5.50 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr7_-_83670356 | 5.48 |
ENSRNOT00000005584
|
Sybu
|
syntabulin |
| chr6_-_143702033 | 5.48 |
ENSRNOT00000051410
|
AABR07065886.1
|
|
| chr3_+_7884822 | 5.46 |
ENSRNOT00000019157
|
Med27
|
mediator complex subunit 27 |
| chr18_+_30036887 | 5.42 |
ENSRNOT00000077824
|
Pcdha4
|
protocadherin alpha 4 |
| chr17_-_15519060 | 5.34 |
ENSRNOT00000093624
|
Aspnl1
|
asporin-like 1 |
| chr18_+_29987206 | 5.33 |
ENSRNOT00000027383
|
Pcdha4
|
protocadherin alpha 4 |
| chr17_-_52477575 | 5.30 |
ENSRNOT00000081290
|
Gli3
|
GLI family zinc finger 3 |
| chr2_+_202200797 | 5.28 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr6_-_140880070 | 5.27 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr3_-_16441030 | 5.25 |
ENSRNOT00000047784
|
AABR07051532.1
|
|
| chr9_-_30844199 | 5.25 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr3_-_71798531 | 5.24 |
ENSRNOT00000088170
|
Calcrl
|
calcitonin receptor like receptor |
| chr3_+_93909156 | 5.23 |
ENSRNOT00000090365
|
Lmo2
|
LIM domain only 2 |
| chr4_-_119327822 | 5.20 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr1_+_229416489 | 5.20 |
ENSRNOT00000028617
|
Olr336
|
olfactory receptor 336 |
| chr15_-_93868292 | 5.16 |
ENSRNOT00000093546
ENSRNOT00000014583 |
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr1_+_74251167 | 5.16 |
ENSRNOT00000040441
|
Vom2r29
|
vomeronasal 2 receptor, 29 |
| chr4_+_102489916 | 5.10 |
ENSRNOT00000082031
|
AABR07061001.1
|
|
| chr4_-_119131202 | 5.08 |
ENSRNOT00000011675
|
Antxr1
|
anthrax toxin receptor 1 |
| chrX_+_14019961 | 5.07 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr3_+_44025300 | 5.06 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr16_-_28716885 | 5.05 |
ENSRNOT00000059750
|
Spock3
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 3 |
| chr7_-_107223047 | 5.04 |
ENSRNOT00000007250
ENSRNOT00000084875 |
Lrrc6
|
leucine rich repeat containing 6 |
| chr13_+_6679368 | 5.04 |
ENSRNOT00000067198
ENSRNOT00000092965 |
Cntnap5c
|
contactin associated protein-like 5C |
| chr13_+_27312498 | 5.04 |
ENSRNOT00000003466
|
Serpinb7
|
serpin family B member 7 |
| chr2_+_86891092 | 5.02 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr1_+_252409268 | 5.01 |
ENSRNOT00000026219
|
Lipm
|
lipase, family member M |
| chr13_-_27192592 | 5.00 |
ENSRNOT00000040021
|
Serpinb3
|
serpin family B member 3 |
| chr4_+_156253079 | 4.98 |
ENSRNOT00000013536
|
Clec4d
|
C-type lectin domain family 4, member D |
| chr2_+_187447501 | 4.95 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr10_-_110232843 | 4.92 |
ENSRNOT00000054934
|
Cd7
|
Cd7 molecule |
| chr1_-_66237501 | 4.88 |
ENSRNOT00000073006
|
Zfp606
|
zinc finger protein 606 |
| chr10_-_55560422 | 4.80 |
ENSRNOT00000006883
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr1_-_67065797 | 4.79 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chrX_+_110016995 | 4.77 |
ENSRNOT00000093542
|
Nrk
|
Nik related kinase |
| chrX_+_29504349 | 4.73 |
ENSRNOT00000005981
|
Tceanc
|
transcription elongation factor A N-terminal and central domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of Alx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.9 | 44.6 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 7.9 | 23.7 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 5.9 | 17.8 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 5.2 | 20.9 | GO:2001288 | detection of muscle stretch(GO:0035995) positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 5.2 | 31.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 4.6 | 13.7 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 4.5 | 17.9 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 3.7 | 29.5 | GO:0015884 | folic acid transport(GO:0015884) |
| 3.6 | 10.7 | GO:0045399 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 3.3 | 10.0 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 3.2 | 9.5 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 3.1 | 3.1 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) |
| 3.1 | 15.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 3.0 | 14.9 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 3.0 | 8.9 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 2.8 | 22.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 2.7 | 10.7 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 2.6 | 7.9 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 2.5 | 12.7 | GO:0060005 | vestibular reflex(GO:0060005) |
| 2.5 | 7.5 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 2.4 | 7.2 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 2.4 | 9.5 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 2.4 | 7.1 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 2.3 | 9.3 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 2.3 | 6.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 2.3 | 9.0 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 2.1 | 8.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 2.0 | 7.9 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 2.0 | 9.9 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 1.9 | 7.7 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 1.9 | 5.8 | GO:0016598 | protein arginylation(GO:0016598) |
| 1.9 | 5.6 | GO:0097213 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 1.7 | 5.0 | GO:0035425 | autocrine signaling(GO:0035425) |
| 1.5 | 4.5 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.5 | 4.5 | GO:2000771 | epidermal stem cell homeostasis(GO:0036334) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 1.5 | 8.9 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 1.4 | 5.7 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.4 | 12.8 | GO:0070417 | cellular response to cold(GO:0070417) |
| 1.3 | 4.0 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 1.3 | 8.0 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 1.3 | 5.3 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 1.3 | 5.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.3 | 11.7 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 1.3 | 5.0 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.2 | 6.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 1.2 | 3.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 1.2 | 15.0 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 1.1 | 3.4 | GO:0045799 | MAPK import into nucleus(GO:0000189) regulation of chromatin assembly(GO:0010847) positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 1.1 | 4.5 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 1.1 | 5.6 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 1.1 | 17.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 1.1 | 3.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 1.1 | 7.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 1.0 | 4.2 | GO:1903015 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.0 | 3.1 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 1.0 | 5.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 1.0 | 10.8 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 1.0 | 5.9 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.9 | 2.8 | GO:0043474 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 0.9 | 4.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.9 | 10.8 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.9 | 16.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.9 | 22.9 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.9 | 2.6 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.9 | 6.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.9 | 19.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.9 | 2.6 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.8 | 10.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.8 | 2.5 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.8 | 4.0 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.8 | 16.2 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.8 | 4.8 | GO:0060723 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.8 | 11.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.8 | 4.6 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.8 | 2.3 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.8 | 1.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.8 | 9.1 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.8 | 6.8 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.7 | 7.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.7 | 7.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.7 | 4.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.7 | 2.8 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.7 | 7.0 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.7 | 2.7 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.7 | 3.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.6 | 2.5 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.6 | 16.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.6 | 4.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.6 | 7.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.6 | 4.8 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.6 | 6.5 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.6 | 4.6 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.6 | 2.9 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.6 | 5.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.6 | 2.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.5 | 3.8 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.5 | 2.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.5 | 6.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.5 | 3.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.5 | 4.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.5 | 1.5 | GO:0071245 | cellular response to carbon monoxide(GO:0071245) cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) |
| 0.5 | 6.8 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.5 | 1.9 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.5 | 4.3 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.5 | 1.4 | GO:2000464 | positive regulation of astrocyte chemotaxis(GO:2000464) |
| 0.5 | 1.9 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.5 | 3.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.5 | 3.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.5 | 1.9 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.5 | 2.8 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.4 | 3.6 | GO:0033504 | floor plate development(GO:0033504) |
| 0.4 | 3.5 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.4 | 1.8 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.4 | 7.5 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.4 | 2.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 5.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.4 | 4.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.4 | 1.3 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.4 | 1.3 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.4 | 2.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.4 | 5.0 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.4 | 5.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 1.4 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.3 | 0.7 | GO:0000958 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.3 | 7.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.3 | 3.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) regulation of histone phosphorylation(GO:0033127) |
| 0.3 | 3.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.3 | 1.0 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.3 | 0.3 | GO:2000292 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.3 | 11.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.3 | 5.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 1.9 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.3 | 7.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 1.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.3 | 3.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 2.0 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.3 | 1.7 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 11.5 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.3 | 6.7 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.3 | 31.1 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.3 | 8.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 | 3.8 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.3 | 11.7 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 | 7.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 8.4 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.2 | 1.7 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 3.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 1.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 1.8 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.2 | 0.9 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 8.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 1.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 3.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.2 | 3.8 | GO:0050716 | regulation of interleukin-1 beta secretion(GO:0050706) positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.2 | 1.0 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.2 | 3.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 0.6 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.2 | 1.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 2.3 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.2 | 0.4 | GO:0072319 | late endosomal microautophagy(GO:0061738) vesicle uncoating(GO:0072319) |
| 0.2 | 1.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.2 | 1.5 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.2 | 0.9 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 4.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 | 3.5 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 2.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 1.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 0.7 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.2 | 10.2 | GO:0070228 | regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.2 | 1.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 3.2 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.2 | 23.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 3.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.2 | 0.3 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.2 | 0.6 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.2 | 3.9 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.2 | 1.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 4.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 4.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 10.5 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 13.5 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 1.7 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 1.4 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 6.1 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 5.9 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 6.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 8.6 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.1 | 2.1 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 1.0 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 1.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 5.3 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.1 | 0.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 3.4 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.1 | 7.0 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 8.4 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 3.6 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 2.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.9 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.6 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 1.0 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 4.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 6.1 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.1 | 2.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 2.8 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 2.2 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 2.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 5.2 | GO:0030198 | extracellular matrix organization(GO:0030198) |
| 0.1 | 1.2 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.1 | 6.2 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 1.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.5 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 1.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.9 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.1 | 6.8 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.1 | 2.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 3.2 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 5.9 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 0.4 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.7 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 2.4 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 2.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 2.4 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 6.4 | GO:0051403 | stress-activated MAPK cascade(GO:0051403) |
| 0.0 | 0.9 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.8 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 2.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 3.3 | GO:0071222 | cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 1.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 3.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 5.3 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 2.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.0 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 1.3 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 2.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 1.0 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 1.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 6.8 | GO:0007155 | cell adhesion(GO:0007155) |
| 0.0 | 5.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.2 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.3 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.7 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 1.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 31.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 7.4 | 44.6 | GO:0032280 | symmetric synapse(GO:0032280) |
| 4.5 | 17.8 | GO:0044299 | C-fiber(GO:0044299) |
| 3.6 | 10.7 | GO:0097444 | spine apparatus(GO:0097444) |
| 3.0 | 9.0 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 2.6 | 7.9 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 2.2 | 35.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 2.1 | 12.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 2.1 | 6.2 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 1.9 | 13.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 1.9 | 5.6 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 1.8 | 34.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 1.8 | 19.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 1.6 | 6.5 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 1.5 | 4.5 | GO:0060187 | cell pole(GO:0060187) |
| 1.5 | 5.9 | GO:0044307 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 1.4 | 8.6 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.4 | 15.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 1.3 | 10.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.2 | 3.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.1 | 5.3 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 1.0 | 4.1 | GO:0097196 | Shu complex(GO:0097196) |
| 1.0 | 4.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.9 | 4.6 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.9 | 4.5 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.9 | 4.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.8 | 3.9 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.6 | 3.8 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.6 | 6.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.6 | 1.7 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.6 | 2.9 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.5 | 5.5 | GO:0097433 | dense body(GO:0097433) |
| 0.5 | 3.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.5 | 6.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.5 | 7.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.5 | 8.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.5 | 6.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.5 | 6.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.4 | 8.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 3.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.4 | 6.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 22.2 | GO:0031672 | A band(GO:0031672) |
| 0.3 | 4.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 25.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.3 | 1.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 3.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.3 | 4.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 2.9 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.3 | 1.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.3 | 3.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.3 | 5.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.3 | 14.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 21.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.3 | 1.8 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.3 | 5.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.3 | 3.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 1.7 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.3 | 7.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 4.6 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 2.1 | GO:0033290 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 9.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 1.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.3 | 2.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 2.6 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.3 | 1.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 1.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 4.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 4.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.3 | 1.5 | GO:0071547 | piP-body(GO:0071547) |
| 0.2 | 12.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 7.9 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 0.7 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 6.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 1.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 0.7 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.2 | 23.6 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 1.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 7.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 22.5 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 14.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 1.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 2.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 1.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 3.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 3.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 9.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 6.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 7.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 4.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 18.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 5.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 3.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 5.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 7.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.3 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 6.8 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 0.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 23.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 4.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 5.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 10.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 19.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 3.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 9.5 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.1 | 2.6 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.1 | 14.1 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 15.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 29.0 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 31.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 18.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 8.7 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.8 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 1.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 4.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.7 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 23.7 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 7.8 | 31.1 | GO:0019002 | GMP binding(GO:0019002) |
| 7.4 | 44.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 3.7 | 14.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 3.7 | 29.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 3.2 | 19.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 3.0 | 23.8 | GO:0071253 | connexin binding(GO:0071253) |
| 2.6 | 10.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 2.4 | 7.1 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 2.1 | 6.2 | GO:0042498 | diacyl lipopeptide binding(GO:0042498) |
| 2.0 | 10.0 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 2.0 | 35.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.9 | 7.6 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 1.8 | 10.7 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 1.7 | 5.2 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 1.6 | 4.9 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 1.6 | 8.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 1.5 | 4.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 1.4 | 4.3 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 1.4 | 5.6 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 1.4 | 6.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.4 | 2.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.2 | 4.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.1 | 7.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.1 | 7.7 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 1.0 | 16.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.0 | 4.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.0 | 7.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.0 | 5.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.0 | 6.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.9 | 4.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.9 | 3.5 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.9 | 2.6 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.9 | 4.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.9 | 4.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.9 | 11.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.8 | 22.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.8 | 6.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.8 | 6.7 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.7 | 9.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.7 | 5.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.7 | 7.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.6 | 2.6 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.6 | 1.9 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.6 | 4.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.6 | 7.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.6 | 7.6 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.5 | 7.9 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.5 | 1.5 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.5 | 11.7 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.5 | 11.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.5 | 11.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.5 | 3.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.5 | 8.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.5 | 3.9 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.5 | 2.8 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.5 | 5.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 12.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 9.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.4 | 9.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.4 | 5.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 3.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 3.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 1.3 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.4 | 1.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.4 | 1.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.4 | 15.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.4 | 42.1 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.3 | 4.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 33.9 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.3 | 7.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.3 | 3.8 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.3 | 7.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.3 | 1.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.3 | 1.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 3.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.3 | 5.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.3 | 1.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 2.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 1.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 5.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 15.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 2.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 3.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 8.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.2 | 0.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 2.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 12.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 6.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 1.0 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 1.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 8.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 2.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 4.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 4.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 0.9 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 6.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 17.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 5.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 9.6 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 39.6 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 6.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 1.9 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 2.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 8.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 3.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 3.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 6.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 1.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 2.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 4.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 3.1 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 37.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 7.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 26.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 23.8 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 0.6 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.3 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 1.6 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 3.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.9 | GO:0070035 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 2.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 2.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 2.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 7.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 2.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 5.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 14.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 5.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.3 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.2 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.2 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 16.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.9 | 20.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.8 | 9.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.8 | 11.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.8 | 64.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.5 | 8.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 16.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.4 | 8.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.4 | 25.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.4 | 11.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.4 | 6.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 10.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 7.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.3 | 8.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 10.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 11.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.3 | 8.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 9.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 5.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 3.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 6.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 3.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 7.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 6.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 8.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 5.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 3.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 26.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 2.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.2 | 3.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 4.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 9.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 2.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 9.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.6 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 4.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 4.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 8.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 6.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 4.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 2.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 2.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 13.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 1.3 | 10.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.9 | 15.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.8 | 4.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.8 | 9.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.7 | 9.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.6 | 6.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.6 | 6.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.6 | 5.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.6 | 8.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.5 | 31.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.5 | 11.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.5 | 6.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.5 | 18.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.5 | 21.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 6.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.4 | 4.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 3.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 8.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 4.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 7.9 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.3 | 4.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 4.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 7.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 14.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 1.9 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.2 | 5.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 9.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 3.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 4.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 10.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 4.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 2.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 3.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 3.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 1.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 2.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 3.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 5.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.2 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.1 | 8.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.9 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 16.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.0 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 3.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 1.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 3.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 8.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 5.8 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.1 | 8.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 9.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 7.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 1.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 3.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 6.8 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 8.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 4.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 6.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |